Please be patient as the page loads
|
PLS4_MOUSE
|
||||||
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLS4_MOUSE
|
||||||
| θ value | 5.9214e-177 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
PLS4_HUMAN
|
||||||
| θ value | 1.33145e-144 (rank : 2) | NC score | 0.991987 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS2_MOUSE
|
||||||
| θ value | 1.83746e-69 (rank : 3) | NC score | 0.963925 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |
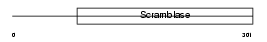
|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_MOUSE
|
||||||
| θ value | 5.17823e-64 (rank : 4) | NC score | 0.951345 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS1_HUMAN
|
||||||
| θ value | 2.03627e-60 (rank : 5) | NC score | 0.959362 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS2_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 6) | NC score | 0.960740 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS3_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 7) | NC score | 0.936982 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |
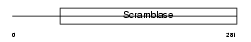
|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 8) | NC score | 0.930915 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.029945 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.050262 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.040409 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.025144 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DLX5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.011961 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
DLX5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.011933 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
ANTR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.027056 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |
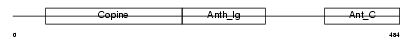
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.026629 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
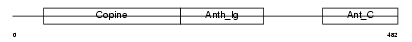
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.026718 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.026790 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.022840 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ZN575_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.000149 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
PER1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.010468 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.014276 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.020378 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.080315 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.080201 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.008400 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
FAIM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.016302 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K097, Q3TY22, Q8K1F6, Q9D6K4 | Gene names | Faim2, Kiaa0950, Lfg | |||
|
Domain Architecture |
|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.007873 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.015925 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.014298 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TIEG3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.000097 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1S5, Q8BI37 | Gene names | Tieg3, Tieg2b | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-inducible early growth response protein 3 (TGFB-inducible early growth response protein 3) (TIEG-3) (TGFB-inducible early growth response protein 2b). | |||||
|
FAIM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.014722 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BWQ8, Q9UJY9, Q9Y2F7 | Gene names | FAIM2, KIAA0950, LFG, TMBIM2 | |||
|
Domain Architecture |
|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein) (Transmembrane BAX inhibitor motif-containing protein 2). | |||||
|
PROP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.005287 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75360 | Gene names | PROP1 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein prophet of Pit-1 (PROP-1) (Pituitary-specific homeodomain factor). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.008247 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
PLS4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.9214e-177 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.991987 (rank : 2) | θ value | 1.33145e-144 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS2_MOUSE
|
||||||
| NC score | 0.963925 (rank : 3) | θ value | 1.83746e-69 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS2_HUMAN
|
||||||
| NC score | 0.960740 (rank : 4) | θ value | 9.16162e-53 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_HUMAN
|
||||||
| NC score | 0.959362 (rank : 5) | θ value | 2.03627e-60 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS1_MOUSE
|
||||||
| NC score | 0.951345 (rank : 6) | θ value | 5.17823e-64 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS3_HUMAN
|
||||||
| NC score | 0.936982 (rank : 7) | θ value | 1.85029e-45 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.930915 (rank : 8) | θ value | 1.05066e-40 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.080315 (rank : 9) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.080201 (rank : 10) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.050262 (rank : 11) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.040409 (rank : 12) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.029945 (rank : 13) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
ANTR1_HUMAN
|
||||||
| NC score | 0.027056 (rank : 14) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.026790 (rank : 15) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.026718 (rank : 16) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.026629 (rank : 17) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.025144 (rank : 18) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.022840 (rank : 19) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.020378 (rank : 20) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
FAIM2_MOUSE
|
||||||
| NC score | 0.016302 (rank : 21) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K097, Q3TY22, Q8K1F6, Q9D6K4 | Gene names | Faim2, Kiaa0950, Lfg | |||
|
Domain Architecture |

|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.015925 (rank : 22) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
FAIM2_HUMAN
|
||||||
| NC score | 0.014722 (rank : 23) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BWQ8, Q9UJY9, Q9Y2F7 | Gene names | FAIM2, KIAA0950, LFG, TMBIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein) (Transmembrane BAX inhibitor motif-containing protein 2). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.014298 (rank : 24) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.014276 (rank : 25) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
DLX5_HUMAN
|
||||||
| NC score | 0.011961 (rank : 26) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
DLX5_MOUSE
|
||||||
| NC score | 0.011933 (rank : 27) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.010468 (rank : 28) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.008400 (rank : 29) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.008247 (rank : 30) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.007873 (rank : 31) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
PROP1_HUMAN
|
||||||
| NC score | 0.005287 (rank : 32) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75360 | Gene names | PROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein prophet of Pit-1 (PROP-1) (Pituitary-specific homeodomain factor). | |||||
|
ZN575_HUMAN
|
||||||
| NC score | 0.000149 (rank : 33) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
TIEG3_MOUSE
|
||||||
| NC score | 0.000097 (rank : 34) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1S5, Q8BI37 | Gene names | Tieg3, Tieg2b | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-inducible early growth response protein 3 (TGFB-inducible early growth response protein 3) (TIEG-3) (TGFB-inducible early growth response protein 2b). | |||||