Please be patient as the page loads
|
PLS2_HUMAN
|
||||||
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLS2_HUMAN
|
||||||
| θ value | 8.71062e-112 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS2_MOUSE
|
||||||
| θ value | 6.47494e-99 (rank : 2) | NC score | 0.995449 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |
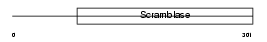
|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_HUMAN
|
||||||
| θ value | 1.88391e-98 (rank : 3) | NC score | 0.995559 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS1_MOUSE
|
||||||
| θ value | 1.39714e-93 (rank : 4) | NC score | 0.982391 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS3_HUMAN
|
||||||
| θ value | 2.03155e-68 (rank : 5) | NC score | 0.978945 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |
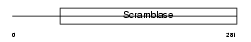
|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 3.14151e-61 (rank : 6) | NC score | 0.971345 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS4_HUMAN
|
||||||
| θ value | 2.84797e-54 (rank : 7) | NC score | 0.962471 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS4_MOUSE
|
||||||
| θ value | 9.16162e-53 (rank : 8) | NC score | 0.960740 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 9) | NC score | 0.053908 (rank : 9) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.053833 (rank : 10) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
PLS2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.71062e-112 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_HUMAN
|
||||||
| NC score | 0.995559 (rank : 2) | θ value | 1.88391e-98 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS2_MOUSE
|
||||||
| NC score | 0.995449 (rank : 3) | θ value | 6.47494e-99 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_MOUSE
|
||||||
| NC score | 0.982391 (rank : 4) | θ value | 1.39714e-93 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS3_HUMAN
|
||||||
| NC score | 0.978945 (rank : 5) | θ value | 2.03155e-68 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.971345 (rank : 6) | θ value | 3.14151e-61 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.962471 (rank : 7) | θ value | 2.84797e-54 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS4_MOUSE
|
||||||
| NC score | 0.960740 (rank : 8) | θ value | 9.16162e-53 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.053908 (rank : 9) | θ value | θ > 10 (rank : 9) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.053833 (rank : 10) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||