Please be patient as the page loads
|
PLS4_HUMAN
|
||||||
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLS4_HUMAN
|
||||||
| θ value | 7.01099e-170 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS4_MOUSE
|
||||||
| θ value | 1.33145e-144 (rank : 2) | NC score | 0.991987 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
PLS2_MOUSE
|
||||||
| θ value | 1.27248e-70 (rank : 3) | NC score | 0.964534 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |
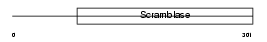
|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_MOUSE
|
||||||
| θ value | 2.10232e-65 (rank : 4) | NC score | 0.953481 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS1_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 5) | NC score | 0.961784 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS2_HUMAN
|
||||||
| θ value | 2.84797e-54 (rank : 6) | NC score | 0.962471 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS3_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 7) | NC score | 0.941087 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |
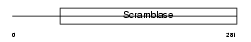
|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 8) | NC score | 0.936414 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.109222 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.109062 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.006615 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.020230 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.034909 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.018239 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.041933 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CXA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.010260 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64448 | Gene names | Gja3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.025912 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
SIP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.004597 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.019889 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.022270 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.032084 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.002019 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
NUFP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.020818 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.033381 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.032851 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.016362 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.004086 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
LMBL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.008341 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JM7, Q5VUM9, Q6P9B5 | Gene names | L3MBTL3, KIAA1798 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein) (H-l(3)mbt-like protein). | |||||
|
PO210_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.009102 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY81, Q3U031, Q69ZW2, Q7TQM1 | Gene names | Nup210, Kiaa0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
SSDH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.005702 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.012851 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.012954 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.017472 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ZN414_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.008074 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCK4, Q8CHU9 | Gene names | Znf414, Zfp414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.01099e-170 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS4_MOUSE
|
||||||
| NC score | 0.991987 (rank : 2) | θ value | 1.33145e-144 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
PLS2_MOUSE
|
||||||
| NC score | 0.964534 (rank : 3) | θ value | 1.27248e-70 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS2_HUMAN
|
||||||
| NC score | 0.962471 (rank : 4) | θ value | 2.84797e-54 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_HUMAN
|
||||||
| NC score | 0.961784 (rank : 5) | θ value | 5.17823e-64 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS1_MOUSE
|
||||||
| NC score | 0.953481 (rank : 6) | θ value | 2.10232e-65 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS3_HUMAN
|
||||||
| NC score | 0.941087 (rank : 7) | θ value | 1.51363e-47 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.936414 (rank : 8) | θ value | 1.19932e-44 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.109222 (rank : 9) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.109062 (rank : 10) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.041933 (rank : 11) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.034909 (rank : 12) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.033381 (rank : 13) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.032851 (rank : 14) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.032084 (rank : 15) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.025912 (rank : 16) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.022270 (rank : 17) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
NUFP1_HUMAN
|
||||||
| NC score | 0.020818 (rank : 18) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.020230 (rank : 19) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.019889 (rank : 20) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.018239 (rank : 21) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.017472 (rank : 22) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.016362 (rank : 23) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.012954 (rank : 24) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.012851 (rank : 25) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CXA3_MOUSE
|
||||||
| NC score | 0.010260 (rank : 26) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64448 | Gene names | Gja3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
PO210_MOUSE
|
||||||
| NC score | 0.009102 (rank : 27) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY81, Q3U031, Q69ZW2, Q7TQM1 | Gene names | Nup210, Kiaa0906 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore membrane glycoprotein 210 precursor (POM210) (Nuclear pore protein gp210). | |||||
|
LMBL3_HUMAN
|
||||||
| NC score | 0.008341 (rank : 28) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JM7, Q5VUM9, Q6P9B5 | Gene names | L3MBTL3, KIAA1798 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein) (H-l(3)mbt-like protein). | |||||
|
ZN414_MOUSE
|
||||||
| NC score | 0.008074 (rank : 29) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCK4, Q8CHU9 | Gene names | Znf414, Zfp414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.006615 (rank : 30) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
SSDH_MOUSE
|
||||||
| NC score | 0.005702 (rank : 31) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
SIP1_MOUSE
|
||||||
| NC score | 0.004597 (rank : 32) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.004086 (rank : 33) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.002019 (rank : 34) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||