Please be patient as the page loads
|
SSDH_MOUSE
|
||||||
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SSDH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998954 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51649 | Gene names | ALDH5A1, SSADH | |||
|
Domain Architecture |

|
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
SSDH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
AL1A2_HUMAN
|
||||||
| θ value | 1.89707e-74 (rank : 3) | NC score | 0.964632 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94788, Q2PJS6, Q8NHQ4, Q9UBR8, Q9UFY0 | Gene names | ALDH1A2, RALDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A2_MOUSE
|
||||||
| θ value | 5.51964e-74 (rank : 4) | NC score | 0.964591 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62148, Q6DI79 | Gene names | Aldh1a2, Aldh1a7, Raldh2 | |||
|
Domain Architecture |
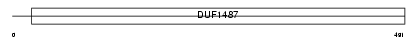
|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
FTHFD_MOUSE
|
||||||
| θ value | 8.81223e-72 (rank : 5) | NC score | 0.954192 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R0Y6 | Gene names | Aldh1l1, Fthfd | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
AL9A1_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 6) | NC score | 0.973150 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLJ2 | Gene names | Aldh9a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3). | |||||
|
AL1A1_HUMAN
|
||||||
| θ value | 1.66191e-70 (rank : 7) | NC score | 0.964365 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00352, O00768 | Gene names | ALDH1A1, ALDC, ALDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1A1_MOUSE
|
||||||
| θ value | 2.83479e-70 (rank : 8) | NC score | 0.965367 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P24549, Q7TQJ0, Q811J0 | Gene names | Aldh1a1, Ahd-2, Ahd2, Aldh1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1A3_HUMAN
|
||||||
| θ value | 2.83479e-70 (rank : 9) | NC score | 0.965250 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47895, Q6NT64 | Gene names | ALDH1A3, ALDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
AL1A3_MOUSE
|
||||||
| θ value | 1.40689e-69 (rank : 10) | NC score | 0.965394 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHW9, Q9EQP7, Q9JI72 | Gene names | Aldh1a3, Aldh6, Raldh3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
FTHFD_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 11) | NC score | 0.954733 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75891, Q68CS1 | Gene names | ALDH1L1, FTHFD | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
AL9A1_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 12) | NC score | 0.972757 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49189, Q5VV90, Q6LCL1, Q9NZT7 | Gene names | ALDH9A1, ALDH7, ALDH9 | |||
|
Domain Architecture |

|
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3) (Aldehyde dehydrogenase E3 isozyme) (Gamma-aminobutyraldehyde dehydrogenase) (EC 1.2.1.19) (R- aminobutyraldehyde dehydrogenase). | |||||
|
ALDH2_HUMAN
|
||||||
| θ value | 2.74572e-65 (rank : 13) | NC score | 0.961890 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05091, Q03639, Q6IB13, Q6IV71 | Gene names | ALDH2, ALDM | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2). | |||||
|
AL1B1_HUMAN
|
||||||
| θ value | 6.32992e-62 (rank : 14) | NC score | 0.962659 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30837 | Gene names | ALDH1B1, ALDH5, ALDHX | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase X, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2). | |||||
|
ALDH2_MOUSE
|
||||||
| θ value | 1.84173e-61 (rank : 15) | NC score | 0.960481 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47738 | Gene names | Aldh2, Ahd-1, Ahd1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2). | |||||
|
MMSA_HUMAN
|
||||||
| θ value | 3.2585e-50 (rank : 16) | NC score | 0.965149 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02252, Q9UKM8 | Gene names | ALDH6A1, MMSDH | |||
|
Domain Architecture |

|
|||||
| Description | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial precursor (EC 1.2.1.27) (MMSDH) (Malonate-semialdehyde dehydrogenase [acylating]) (EC 1.2.1.18). | |||||
|
AL7A1_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 17) | NC score | 0.943640 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49419, O14619, Q6IPU8, Q9BUL4 | Gene names | ALDH7A1, ATQ1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL7A1_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 18) | NC score | 0.943353 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBF1 | Gene names | Aldh7a1, Ald7a1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL3A1_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 19) | NC score | 0.870240 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47739, Q9R203 | Gene names | Aldh3a1, Ahd-4, Ahd4, Aldh3, Aldh4 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (Dioxin-inducible aldehyde dehydrogenase 3). | |||||
|
AL3A1_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 20) | NC score | 0.870990 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30838, Q9BT37 | Gene names | ALDH3A1, ALDH3 | |||
|
Domain Architecture |
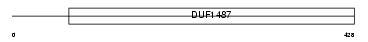
|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (ALDHIII). | |||||
|
AL3B1_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 21) | NC score | 0.852481 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43353, Q53XL5, Q8N515, Q96CK8 | Gene names | ALDH3B1, ALDH7 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
AL3B1_MOUSE
|
||||||
| θ value | 6.38894e-30 (rank : 22) | NC score | 0.853281 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VQ0, Q63ZW3, Q8VHW0, Q9CW05 | Gene names | Aldh3b1, Aldh7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
AL3A2_MOUSE
|
||||||
| θ value | 8.3442e-30 (rank : 23) | NC score | 0.864935 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47740 | Gene names | Aldh3a2, Ahd-3, Ahd3, Aldh3, Aldh4 | |||
|
Domain Architecture |
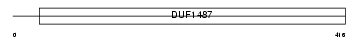
|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3A2_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 24) | NC score | 0.854957 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51648, Q93011, Q96J37 | Gene names | ALDH3A2, ALDH10, FALDH | |||
|
Domain Architecture |

|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3B2_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 25) | NC score | 0.862243 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48448, Q8NAL5, Q96IB2 | Gene names | ALDH3B2, ALDH8 | |||
|
Domain Architecture |
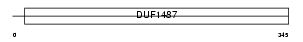
|
|||||
| Description | Aldehyde dehydrogenase 3B2 (EC 1.2.1.5) (Aldehyde dehydrogenase 8). | |||||
|
AL4A1_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 26) | NC score | 0.875250 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30038, Q16882, Q8IZ38, Q96IF0 | Gene names | ALDH4A1, ALDH4, P5CDH | |||
|
Domain Architecture |

|
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL4A1_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 27) | NC score | 0.865376 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHT0, Q7TND0, Q8BXM3, Q8R0N1, Q8R1S2 | Gene names | Aldh4a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
3HIDH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.039544 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31937, Q9UDN3 | Gene names | HIBADH | |||
|
Domain Architecture |
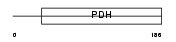
|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
KLF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | -0.001888 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5W3, Q9UJS5, Q9UKR6 | Gene names | KLF2, LKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 2 (Lung krueppel-like factor). | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.014443 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
RS4Y2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.011856 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TD47 | Gene names | RPS4Y2 | |||
|
Domain Architecture |
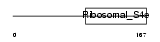
|
|||||
| Description | 40S ribosomal protein S4, Y isoform 2. | |||||
|
3HIDH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.032503 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L13, Q8BJY2 | Gene names | Hibadh | |||
|
Domain Architecture |
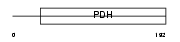
|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
FOXD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.001876 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |
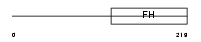
|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.000403 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
DEND_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.009932 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
GRASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.006404 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.010344 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
PLS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.005702 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
RDH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.003212 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TC12, Q9NRW0, Q9Y391 | Gene names | RDH11, ARSDR1, PSDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | -0.002532 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | -0.002681 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
FMT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.071338 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FMT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.074320 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
SSDH_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
SSDH_HUMAN
|
||||||
| NC score | 0.998954 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51649 | Gene names | ALDH5A1, SSADH | |||
|
Domain Architecture |

|
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
AL9A1_MOUSE
|
||||||
| NC score | 0.973150 (rank : 3) | θ value | 3.34864e-71 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLJ2 | Gene names | Aldh9a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3). | |||||
|
AL9A1_HUMAN
|
||||||
| NC score | 0.972757 (rank : 4) | θ value | 7.71989e-68 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49189, Q5VV90, Q6LCL1, Q9NZT7 | Gene names | ALDH9A1, ALDH7, ALDH9 | |||
|
Domain Architecture |

|
|||||
| Description | 4-trimethylaminobutyraldehyde dehydrogenase (EC 1.2.1.47) (TMABADH) (Aldehyde dehydrogenase 9A1) (EC 1.2.1.3) (Aldehyde dehydrogenase E3 isozyme) (Gamma-aminobutyraldehyde dehydrogenase) (EC 1.2.1.19) (R- aminobutyraldehyde dehydrogenase). | |||||
|
AL1A3_MOUSE
|
||||||
| NC score | 0.965394 (rank : 5) | θ value | 1.40689e-69 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHW9, Q9EQP7, Q9JI72 | Gene names | Aldh1a3, Aldh6, Raldh3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
AL1A1_MOUSE
|
||||||
| NC score | 0.965367 (rank : 6) | θ value | 2.83479e-70 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P24549, Q7TQJ0, Q811J0 | Gene names | Aldh1a1, Ahd-2, Ahd2, Aldh1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1A3_HUMAN
|
||||||
| NC score | 0.965250 (rank : 7) | θ value | 2.83479e-70 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47895, Q6NT64 | Gene names | ALDH1A3, ALDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 1A3 (EC 1.2.1.5) (Aldehyde dehydrogenase 6) (Retinaldehyde dehydrogenase 3) (RALDH-3). | |||||
|
MMSA_HUMAN
|
||||||
| NC score | 0.965149 (rank : 8) | θ value | 3.2585e-50 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02252, Q9UKM8 | Gene names | ALDH6A1, MMSDH | |||
|
Domain Architecture |

|
|||||
| Description | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial precursor (EC 1.2.1.27) (MMSDH) (Malonate-semialdehyde dehydrogenase [acylating]) (EC 1.2.1.18). | |||||
|
AL1A2_HUMAN
|
||||||
| NC score | 0.964632 (rank : 9) | θ value | 1.89707e-74 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94788, Q2PJS6, Q8NHQ4, Q9UBR8, Q9UFY0 | Gene names | ALDH1A2, RALDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A2_MOUSE
|
||||||
| NC score | 0.964591 (rank : 10) | θ value | 5.51964e-74 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62148, Q6DI79 | Gene names | Aldh1a2, Aldh1a7, Raldh2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 2 (EC 1.2.1.36) (RalDH2) (RALDH 2) (RALDH(II)) (Retinaldehyde-specific dehydrogenase type 2) (Aldehyde dehydrogenase family 1 member A2). | |||||
|
AL1A1_HUMAN
|
||||||
| NC score | 0.964365 (rank : 11) | θ value | 1.66191e-70 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00352, O00768 | Gene names | ALDH1A1, ALDC, ALDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal dehydrogenase 1 (EC 1.2.1.36) (RalDH1) (RALDH 1) (Aldehyde dehydrogenase family 1 member A1) (Aldehyde dehydrogenase, cytosolic) (ALHDII) (ALDH-E1). | |||||
|
AL1B1_HUMAN
|
||||||
| NC score | 0.962659 (rank : 12) | θ value | 6.32992e-62 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30837 | Gene names | ALDH1B1, ALDH5, ALDHX | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase X, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2). | |||||
|
ALDH2_HUMAN
|
||||||
| NC score | 0.961890 (rank : 13) | θ value | 2.74572e-65 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05091, Q03639, Q6IB13, Q6IV71 | Gene names | ALDH2, ALDM | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2). | |||||
|
ALDH2_MOUSE
|
||||||
| NC score | 0.960481 (rank : 14) | θ value | 1.84173e-61 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47738 | Gene names | Aldh2, Ahd-1, Ahd1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2). | |||||
|
FTHFD_HUMAN
|
||||||
| NC score | 0.954733 (rank : 15) | θ value | 3.13423e-69 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75891, Q68CS1 | Gene names | ALDH1L1, FTHFD | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
FTHFD_MOUSE
|
||||||
| NC score | 0.954192 (rank : 16) | θ value | 8.81223e-72 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R0Y6 | Gene names | Aldh1l1, Fthfd | |||
|
Domain Architecture |

|
|||||
| Description | 10-formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) (10-FTHFDH) (Aldehyde dehydrogenase 1 family member L1). | |||||
|
AL7A1_HUMAN
|
||||||
| NC score | 0.943640 (rank : 17) | θ value | 3.15612e-45 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49419, O14619, Q6IPU8, Q9BUL4 | Gene names | ALDH7A1, ATQ1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL7A1_MOUSE
|
||||||
| NC score | 0.943353 (rank : 18) | θ value | 1.19932e-44 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBF1 | Gene names | Aldh7a1, Ald7a1 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase family 7 member A1 (EC 1.2.1.3) (Antiquitin-1). | |||||
|
AL4A1_HUMAN
|
||||||
| NC score | 0.875250 (rank : 19) | θ value | 1.62847e-25 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30038, Q16882, Q8IZ38, Q96IF0 | Gene names | ALDH4A1, ALDH4, P5CDH | |||
|
Domain Architecture |

|
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL3A1_HUMAN
|
||||||
| NC score | 0.870990 (rank : 20) | θ value | 4.42448e-31 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30838, Q9BT37 | Gene names | ALDH3A1, ALDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (ALDHIII). | |||||
|
AL3A1_MOUSE
|
||||||
| NC score | 0.870240 (rank : 21) | θ value | 4.00176e-32 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47739, Q9R203 | Gene names | Aldh3a1, Ahd-4, Ahd4, Aldh3, Aldh4 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase, dimeric NADP-preferring (EC 1.2.1.5) (ALDH class 3) (Dioxin-inducible aldehyde dehydrogenase 3). | |||||
|
AL4A1_MOUSE
|
||||||
| NC score | 0.865376 (rank : 22) | θ value | 1.09232e-21 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHT0, Q7TND0, Q8BXM3, Q8R0N1, Q8R1S2 | Gene names | Aldh4a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor (EC 1.5.1.12) (P5C dehydrogenase) (Aldehyde dehydrogenase 4A1). | |||||
|
AL3A2_MOUSE
|
||||||
| NC score | 0.864935 (rank : 23) | θ value | 8.3442e-30 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47740 | Gene names | Aldh3a2, Ahd-3, Ahd3, Aldh3, Aldh4 | |||
|
Domain Architecture |

|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3B2_HUMAN
|
||||||
| NC score | 0.862243 (rank : 24) | θ value | 2.27234e-27 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48448, Q8NAL5, Q96IB2 | Gene names | ALDH3B2, ALDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B2 (EC 1.2.1.5) (Aldehyde dehydrogenase 8). | |||||
|
AL3A2_HUMAN
|
||||||
| NC score | 0.854957 (rank : 25) | θ value | 2.05525e-28 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51648, Q93011, Q96J37 | Gene names | ALDH3A2, ALDH10, FALDH | |||
|
Domain Architecture |

|
|||||
| Description | Fatty aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase, microsomal) (Aldehyde dehydrogenase family 3 member A2) (Aldehyde dehydrogenase 10). | |||||
|
AL3B1_MOUSE
|
||||||
| NC score | 0.853281 (rank : 26) | θ value | 6.38894e-30 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VQ0, Q63ZW3, Q8VHW0, Q9CW05 | Gene names | Aldh3b1, Aldh7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
AL3B1_HUMAN
|
||||||
| NC score | 0.852481 (rank : 27) | θ value | 4.89182e-30 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43353, Q53XL5, Q8N515, Q96CK8 | Gene names | ALDH3B1, ALDH7 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B1 (EC 1.2.1.5) (Aldehyde dehydrogenase 7). | |||||
|
FMT_MOUSE
|
||||||
| NC score | 0.074320 (rank : 28) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D799, Q8VE89 | Gene names | Mtfmt, Fmt | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
FMT_HUMAN
|
||||||
| NC score | 0.071338 (rank : 29) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DP5 | Gene names | MTFMT, FMT, FMT1 | |||
|
Domain Architecture |
|
|||||
| Description | Methionyl-tRNA formyltransferase, mitochondrial precursor (EC 2.1.2.9) (MtFMT). | |||||
|
3HIDH_HUMAN
|
||||||
| NC score | 0.039544 (rank : 30) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31937, Q9UDN3 | Gene names | HIBADH | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
3HIDH_MOUSE
|
||||||
| NC score | 0.032503 (rank : 31) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L13, Q8BJY2 | Gene names | Hibadh | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.014443 (rank : 32) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
RS4Y2_HUMAN
|
||||||
| NC score | 0.011856 (rank : 33) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TD47 | Gene names | RPS4Y2 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein S4, Y isoform 2. | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.010344 (rank : 34) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.009932 (rank : 35) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
GRASP_HUMAN
|
||||||
| NC score | 0.006404 (rank : 36) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.005702 (rank : 37) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
RDH11_HUMAN
|
||||||
| NC score | 0.003212 (rank : 38) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TC12, Q9NRW0, Q9Y391 | Gene names | RDH11, ARSDR1, PSDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12). | |||||
|
FOXD2_HUMAN
|
||||||
| NC score | 0.001876 (rank : 39) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.000403 (rank : 40) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
KLF2_HUMAN
|
||||||
| NC score | -0.001888 (rank : 41) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5W3, Q9UJS5, Q9UKR6 | Gene names | KLF2, LKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 2 (Lung krueppel-like factor). | |||||
|
K1802_HUMAN
|
||||||
| NC score | -0.002532 (rank : 42) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | -0.002681 (rank : 43) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||