Please be patient as the page loads
|
FZD6_MOUSE
|
||||||
| SwissProt Accessions | Q61089 | Gene names | Fzd6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (mFz6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FZD6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994397 (rank : 2) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60353, Q6P9C3 | Gene names | FZD6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (hFz6). | |||||
|
FZD6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | Q61089 | Gene names | Fzd6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (mFz6). | |||||
|
FZD3_MOUSE
|
||||||
| θ value | 1.71888e-184 (rank : 3) | NC score | 0.987311 (rank : 3) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61086 | Gene names | Fzd3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-3 precursor (Fz-3) (mFz3). | |||||
|
FZD3_HUMAN
|
||||||
| θ value | 8.53067e-184 (rank : 4) | NC score | 0.987245 (rank : 4) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NPG1 | Gene names | FZD3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-3 precursor (Fz-3) (hFz3). | |||||
|
FZD2_MOUSE
|
||||||
| θ value | 1.63515e-126 (rank : 5) | NC score | 0.977209 (rank : 7) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JIP6, Q810M0, Q9JIP5, Q9WUJ2 | Gene names | Fzd2, Fzd10 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (mFz2) (mFz10). | |||||
|
FZD1_HUMAN
|
||||||
| θ value | 2.78914e-126 (rank : 6) | NC score | 0.977726 (rank : 6) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UP38, O94815, Q549T8 | Gene names | FZD1 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-1 precursor (Fz-1) (hFz1) (FzE1). | |||||
|
FZD7_HUMAN
|
||||||
| θ value | 8.11517e-126 (rank : 7) | NC score | 0.976632 (rank : 10) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75084, O94816, Q96B74 | Gene names | FZD7 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-7 precursor (Fz-7) (hFz7) (FzE3). | |||||
|
FZD2_HUMAN
|
||||||
| θ value | 8.97236e-125 (rank : 8) | NC score | 0.977084 (rank : 8) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14332 | Gene names | FZD2 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (hFz2) (FzE2). | |||||
|
FZD7_MOUSE
|
||||||
| θ value | 1.53045e-124 (rank : 9) | NC score | 0.976741 (rank : 9) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61090 | Gene names | Fzd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Frizzled-7 precursor (Fz-7) (mFz7). | |||||
|
FZD1_MOUSE
|
||||||
| θ value | 3.19119e-122 (rank : 10) | NC score | 0.977895 (rank : 5) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O70421, O08974 | Gene names | Fzd1 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-1 precursor (Fz-1) (mFz1). | |||||
|
FZD10_MOUSE
|
||||||
| θ value | 1.39714e-93 (rank : 11) | NC score | 0.966332 (rank : 14) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BKG4 | Gene names | Fzd10, Fz10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Frizzled-10 precursor (Fz-10). | |||||
|
FZD10_HUMAN
|
||||||
| θ value | 4.96919e-91 (rank : 12) | NC score | 0.966522 (rank : 13) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULW2 | Gene names | FZD10 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-10 precursor (Fz-10) (hFz10) (FzE7). | |||||
|
FZD4_HUMAN
|
||||||
| θ value | 4.65103e-89 (rank : 13) | NC score | 0.966765 (rank : 12) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULV1, Q6S9E4 | Gene names | FZD4 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-4 precursor (Fz-4) (hFz4) (FzE4). | |||||
|
FZD4_MOUSE
|
||||||
| θ value | 1.35324e-88 (rank : 14) | NC score | 0.966770 (rank : 11) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61088 | Gene names | Fzd4 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-4 precursor (Fz-4) (mFz4). | |||||
|
FZD9_MOUSE
|
||||||
| θ value | 3.33315e-87 (rank : 15) | NC score | 0.965470 (rank : 16) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R216, O35494, Q9CX16, Q9R2B3 | Gene names | Fzd9, Fzd3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-9 precursor (Fz-9) (mFz9) (mFz3). | |||||
|
FZD9_HUMAN
|
||||||
| θ value | 9.07703e-85 (rank : 16) | NC score | 0.965370 (rank : 18) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00144 | Gene names | FZD9, FZD3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-9 precursor (Fz-9) (hFz9) (FzE6). | |||||
|
FZD5_MOUSE
|
||||||
| θ value | 2.02215e-84 (rank : 17) | NC score | 0.965403 (rank : 17) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9EQD0, O08975, Q8BMR2, Q8CHK9 | Gene names | Fzd5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (mFz5). | |||||
|
FZD5_HUMAN
|
||||||
| θ value | 3.44927e-84 (rank : 18) | NC score | 0.966284 (rank : 15) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13467, Q53R22 | Gene names | FZD5, HFZ5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (hFz5) (FzE5). | |||||
|
FZD8_MOUSE
|
||||||
| θ value | 6.52014e-75 (rank : 19) | NC score | 0.959227 (rank : 19) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61091 | Gene names | Fzd8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (mFz8). | |||||
|
FZD8_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 20) | NC score | 0.958918 (rank : 20) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H461 | Gene names | FZD8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (hFz8). | |||||
|
SMO_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 21) | NC score | 0.890264 (rank : 21) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99835 | Gene names | SMO, SMOH | |||
|
Domain Architecture |

|
|||||
| Description | Smoothened homolog precursor (SMO) (Gx protein). | |||||
|
SMO_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 22) | NC score | 0.886806 (rank : 22) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56726 | Gene names | Smo, Smoh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smoothened homolog precursor (SMO). | |||||
|
SFRP3_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 23) | NC score | 0.823411 (rank : 25) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97401, O09075, O09093, Q91W58 | Gene names | Frzb, Fiz, Fre, Frzb1, Sfrp3 | |||
|
Domain Architecture |
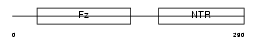
|
|||||
| Description | Secreted frizzled-related protein 3 precursor (sFRP-3) (Frizzled- related protein 1) (FrzB-1) (Frezzled) (Fritz). | |||||
|
SFRP4_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 24) | NC score | 0.833076 (rank : 24) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z1N6, Q91ZX9 | Gene names | Sfrp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related sequence protein 4 precursor (sFRP-4) (Frizzled-related protein 4) (FRP-4). | |||||
|
SFRP3_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 25) | NC score | 0.822057 (rank : 26) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92765, O00181, Q99686 | Gene names | FRZB, FIZ, FRE, FRP, FRZB1, SFRP3 | |||
|
Domain Architecture |
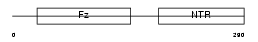
|
|||||
| Description | Secreted frizzled-related protein 3 precursor (sFRP-3) (Frizzled- related protein 1) (FrzB-1) (Frezzled) (Fritz). | |||||
|
SFRP4_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 26) | NC score | 0.833154 (rank : 23) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6FHJ7, O14877, Q4G124, Q6FHM0, Q6PD64 | Gene names | SFRP4, FRPHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 4 precursor (sFRP-4) (Frizzled protein, human endometrium) (FrpHE). | |||||
|
SFRP1_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 27) | NC score | 0.794290 (rank : 27) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C4U3, O08861, Q8R1J4 | Gene names | Sfrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 1 precursor (sFRP-1). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 28) | NC score | 0.278584 (rank : 37) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
SFRP2_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 29) | NC score | 0.788177 (rank : 32) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96HF1, O14778, Q9HAP5 | Gene names | SFRP2, FRP2, SARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (Frizzled-related protein 2) (FRP-2). | |||||
|
SFRP2_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 30) | NC score | 0.789066 (rank : 31) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97299, O08862, O35297 | Gene names | Sfrp2, Sarp1, Sdf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (SDF5 protein). | |||||
|
SFRP1_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 31) | NC score | 0.792919 (rank : 28) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N474, O00546, O14779 | Gene names | SFRP1, FRP, FRP1, SARP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 1 precursor (sFRP-1) (Frizzled- related protein 1) (FRP-1) (Secreted apoptosis-related protein 2) (SARP-2). | |||||
|
CORIN_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 32) | NC score | 0.278168 (rank : 38) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
SFRP5_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 33) | NC score | 0.790280 (rank : 29) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5T4F7, O14780, Q86TH7 | Gene names | SFRP5, FRP1B, SARP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5) (Secreted apoptosis-related protein 3) (SARP-3) (Frizzled-related protein 1b) (FRP-1b). | |||||
|
SFRP5_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 34) | NC score | 0.790067 (rank : 30) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WU66, Q8K269 | Gene names | Sfrp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 35) | NC score | 0.441534 (rank : 33) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 36) | NC score | 0.435046 (rank : 34) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
CBPZ_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 37) | NC score | 0.418081 (rank : 35) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 38) | NC score | 0.373576 (rank : 36) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
BAI1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 39) | NC score | 0.085060 (rank : 47) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 40) | NC score | 0.088477 (rank : 46) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 41) | NC score | 0.094251 (rank : 45) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 42) | NC score | 0.094257 (rank : 44) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 43) | NC score | 0.082809 (rank : 48) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60242, O60297 | Gene names | BAI3, KIAA0550 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
BAI3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 44) | NC score | 0.082624 (rank : 49) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80ZF8 | Gene names | Bai3 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 45) | NC score | 0.243146 (rank : 39) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GP133_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 46) | NC score | 0.097864 (rank : 43) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 47) | NC score | 0.035499 (rank : 60) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 48) | NC score | 0.019234 (rank : 79) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
HORN_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 49) | NC score | 0.033085 (rank : 62) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.024023 (rank : 72) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.016778 (rank : 83) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.028360 (rank : 67) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
GP155_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.147519 (rank : 42) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.025783 (rank : 70) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.018546 (rank : 81) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.021240 (rank : 75) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CECR6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.036182 (rank : 58) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXQ6 | Gene names | CECR6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6. | |||||
|
CECR6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.035643 (rank : 59) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MX7 | Gene names | Cecr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6 homolog. | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.022036 (rank : 74) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | 0.022766 (rank : 73) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.018951 (rank : 80) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.015550 (rank : 88) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SYNPR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.031993 (rank : 63) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGN8, Q8C5J4, Q9DB89 | Gene names | Synpr | |||
|
Domain Architecture |
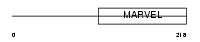
|
|||||
| Description | Synaptoporin. | |||||
|
PEG3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | -0.001743 (rank : 131) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.007415 (rank : 106) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
SYNPR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.031865 (rank : 64) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBG9 | Gene names | SYNPR | |||
|
Domain Architecture |
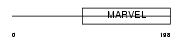
|
|||||
| Description | Synaptoporin. | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.012123 (rank : 94) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.020026 (rank : 77) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.031328 (rank : 65) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.011381 (rank : 96) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.012215 (rank : 93) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.020111 (rank : 76) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.006126 (rank : 110) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
GP157_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.186037 (rank : 40) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5UAW9, Q8WWB8, Q9HA73 | Gene names | GPR157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.016124 (rank : 85) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
TM131_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.018492 (rank : 82) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.007094 (rank : 107) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.007996 (rank : 101) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
GPR98_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.046849 (rank : 56) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.007700 (rank : 105) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
TLE3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.005056 (rank : 118) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.007042 (rank : 108) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
K0226_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.011268 (rank : 97) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
REXO4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.014148 (rank : 89) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR2, Q5T8S4, Q5T8S5, Q5T8S6, Q9GZW3 | Gene names | REXO4, PMC2, XPMC2H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (hPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
S19A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.019825 (rank : 78) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41440, O00553, O60227, Q13026 | Gene names | SLC19A1, FLOT1, RFC1 | |||
|
Domain Architecture |

|
|||||
| Description | Folate transporter 1 (Solute carrier family 19 member 1) (Placental folate transporter) (FOLT) (Reduced folate carrier protein) (RFC) (Intestinal folate carrier) (IFC-1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.026231 (rank : 69) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.006874 (rank : 109) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
BASP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.016185 (rank : 84) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CD97_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.026513 (rank : 68) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48960, O00718, O76101, Q8NG72, Q8TBQ7 | Gene names | CD97 | |||
|
Domain Architecture |

|
|||||
| Description | CD97 antigen precursor (Leukocyte antigen CD97). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.030931 (rank : 66) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
GP157_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.175916 (rank : 41) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C206, Q8C224 | Gene names | Gpr157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.061565 (rank : 51) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
IG2AS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.025580 (rank : 71) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6U949, Q7Z712, Q9P2W0 | Gene names | IGF2AS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative insulin-like growth factor 2 antisense gene protein (IGF2-AS) (PEG8/IGF2AS protein). | |||||
|
LPHN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.060377 (rank : 52) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HAR2, O94867, Q9NWK5 | Gene names | LPHN3, KIAA0768, LEC3 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-3 precursor (Calcium-independent alpha-latrotoxin receptor 3) (Lectomedin-3). | |||||
|
M3K10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | -0.005210 (rank : 138) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |
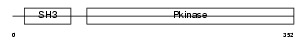
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.013720 (rank : 90) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.013720 (rank : 91) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
RU17_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.008808 (rank : 100) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
TLE3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.004518 (rank : 121) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | -0.002666 (rank : 132) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IQEC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.004667 (rank : 119) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
TLR5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.001891 (rank : 127) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60602, O15456 | Gene names | TLR5, TIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor (Toll/interleukin-1 receptor-like protein 3). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | -0.003190 (rank : 133) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.005517 (rank : 114) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.012607 (rank : 92) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RBM15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.007869 (rank : 104) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.005306 (rank : 117) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.007910 (rank : 103) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.010855 (rank : 98) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.001065 (rank : 130) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.004629 (rank : 120) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TR136_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.004284 (rank : 122) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TQA8, Q7M716 | Gene names | Tas2r136, T2r52, Tas2r36 | |||
|
Domain Architecture |
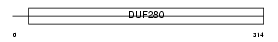
|
|||||
| Description | Taste receptor type 2 member 136 (T2R136) (Taste receptor type 2 member 36) (T2R36) (mT2r52). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | -0.004867 (rank : 137) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.005485 (rank : 115) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
BCAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.009565 (rank : 99) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
GP128_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.036667 (rank : 57) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BM96, Q80T42 | Gene names | Gpr128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
PWWP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.007955 (rank : 102) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.011437 (rank : 95) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.001871 (rank : 128) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.005367 (rank : 116) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SYPL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.015863 (rank : 87) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VXT5, Q6ZMX1 | Gene names | SYPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptophysin-like protein 2. | |||||
|
SYPL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.015883 (rank : 86) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89104 | Gene names | Sypl2, Mg29 | |||
|
Domain Architecture |
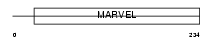
|
|||||
| Description | Synaptophysin-like protein 2 (Mitsugumin-29) (Mg29). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.002977 (rank : 125) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.034370 (rank : 61) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.005832 (rank : 113) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FIBA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.001763 (rank : 129) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.052265 (rank : 55) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
P2RY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | -0.004024 (rank : 134) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47900 | Gene names | P2RY1 | |||
|
Domain Architecture |
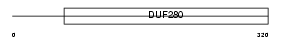
|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.002311 (rank : 126) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.003312 (rank : 124) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SFRS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.005944 (rank : 111) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.005944 (rank : 112) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.003533 (rank : 123) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
ZN440_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | -0.004708 (rank : 136) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYI8, Q8N1R9 | Gene names | ZNF440 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 440. | |||||
|
ZNF24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | -0.004668 (rank : 135) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17028, O14754 | Gene names | ZNF24, KOX17, ZNF191, ZSCAN3 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 24 (Zinc finger protein 191) (Zinc finger protein KOX17) (Retinoic acid suppression protein A) (RSG-A) (Zinc finger and SCAN domain-containing protein 3). | |||||
|
GP112_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.057120 (rank : 53) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP126_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.066704 (rank : 50) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GPR64_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.056120 (rank : 54) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
FZD6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | Q61089 | Gene names | Fzd6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (mFz6). | |||||
|
FZD6_HUMAN
|
||||||
| NC score | 0.994397 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60353, Q6P9C3 | Gene names | FZD6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (hFz6). | |||||
|
FZD3_MOUSE
|
||||||
| NC score | 0.987311 (rank : 3) | θ value | 1.71888e-184 (rank : 3) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61086 | Gene names | Fzd3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-3 precursor (Fz-3) (mFz3). | |||||
|
FZD3_HUMAN
|
||||||
| NC score | 0.987245 (rank : 4) | θ value | 8.53067e-184 (rank : 4) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NPG1 | Gene names | FZD3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-3 precursor (Fz-3) (hFz3). | |||||
|
FZD1_MOUSE
|
||||||
| NC score | 0.977895 (rank : 5) | θ value | 3.19119e-122 (rank : 10) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O70421, O08974 | Gene names | Fzd1 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-1 precursor (Fz-1) (mFz1). | |||||
|
FZD1_HUMAN
|
||||||
| NC score | 0.977726 (rank : 6) | θ value | 2.78914e-126 (rank : 6) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UP38, O94815, Q549T8 | Gene names | FZD1 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-1 precursor (Fz-1) (hFz1) (FzE1). | |||||
|
FZD2_MOUSE
|
||||||
| NC score | 0.977209 (rank : 7) | θ value | 1.63515e-126 (rank : 5) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JIP6, Q810M0, Q9JIP5, Q9WUJ2 | Gene names | Fzd2, Fzd10 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (mFz2) (mFz10). | |||||
|
FZD2_HUMAN
|
||||||
| NC score | 0.977084 (rank : 8) | θ value | 8.97236e-125 (rank : 8) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14332 | Gene names | FZD2 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-2 precursor (Fz-2) (hFz2) (FzE2). | |||||
|
FZD7_MOUSE
|
||||||
| NC score | 0.976741 (rank : 9) | θ value | 1.53045e-124 (rank : 9) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61090 | Gene names | Fzd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Frizzled-7 precursor (Fz-7) (mFz7). | |||||
|
FZD7_HUMAN
|
||||||
| NC score | 0.976632 (rank : 10) | θ value | 8.11517e-126 (rank : 7) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75084, O94816, Q96B74 | Gene names | FZD7 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-7 precursor (Fz-7) (hFz7) (FzE3). | |||||
|
FZD4_MOUSE
|
||||||
| NC score | 0.966770 (rank : 11) | θ value | 1.35324e-88 (rank : 14) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61088 | Gene names | Fzd4 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-4 precursor (Fz-4) (mFz4). | |||||
|
FZD4_HUMAN
|
||||||
| NC score | 0.966765 (rank : 12) | θ value | 4.65103e-89 (rank : 13) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULV1, Q6S9E4 | Gene names | FZD4 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-4 precursor (Fz-4) (hFz4) (FzE4). | |||||
|
FZD10_HUMAN
|
||||||
| NC score | 0.966522 (rank : 13) | θ value | 4.96919e-91 (rank : 12) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULW2 | Gene names | FZD10 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-10 precursor (Fz-10) (hFz10) (FzE7). | |||||
|
FZD10_MOUSE
|
||||||
| NC score | 0.966332 (rank : 14) | θ value | 1.39714e-93 (rank : 11) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BKG4 | Gene names | Fzd10, Fz10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Frizzled-10 precursor (Fz-10). | |||||
|
FZD5_HUMAN
|
||||||
| NC score | 0.966284 (rank : 15) | θ value | 3.44927e-84 (rank : 18) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13467, Q53R22 | Gene names | FZD5, HFZ5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (hFz5) (FzE5). | |||||
|
FZD9_MOUSE
|
||||||
| NC score | 0.965470 (rank : 16) | θ value | 3.33315e-87 (rank : 15) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R216, O35494, Q9CX16, Q9R2B3 | Gene names | Fzd9, Fzd3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-9 precursor (Fz-9) (mFz9) (mFz3). | |||||
|
FZD5_MOUSE
|
||||||
| NC score | 0.965403 (rank : 17) | θ value | 2.02215e-84 (rank : 17) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9EQD0, O08975, Q8BMR2, Q8CHK9 | Gene names | Fzd5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (mFz5). | |||||
|
FZD9_HUMAN
|
||||||
| NC score | 0.965370 (rank : 18) | θ value | 9.07703e-85 (rank : 16) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00144 | Gene names | FZD9, FZD3 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-9 precursor (Fz-9) (hFz9) (FzE6). | |||||
|
FZD8_MOUSE
|
||||||
| NC score | 0.959227 (rank : 19) | θ value | 6.52014e-75 (rank : 19) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61091 | Gene names | Fzd8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (mFz8). | |||||
|
FZD8_HUMAN
|
||||||
| NC score | 0.958918 (rank : 20) | θ value | 3.13423e-69 (rank : 20) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H461 | Gene names | FZD8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (hFz8). | |||||
|
SMO_HUMAN
|
||||||
| NC score | 0.890264 (rank : 21) | θ value | 3.05695e-40 (rank : 21) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99835 | Gene names | SMO, SMOH | |||
|
Domain Architecture |

|
|||||
| Description | Smoothened homolog precursor (SMO) (Gx protein). | |||||
|
SMO_MOUSE
|
||||||
| NC score | 0.886806 (rank : 22) | θ value | 3.37985e-39 (rank : 22) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56726 | Gene names | Smo, Smoh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smoothened homolog precursor (SMO). | |||||
|
SFRP4_HUMAN
|
||||||
| NC score | 0.833154 (rank : 23) | θ value | 8.10077e-17 (rank : 26) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6FHJ7, O14877, Q4G124, Q6FHM0, Q6PD64 | Gene names | SFRP4, FRPHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 4 precursor (sFRP-4) (Frizzled protein, human endometrium) (FrpHE). | |||||
|
SFRP4_MOUSE
|
||||||
| NC score | 0.833076 (rank : 24) | θ value | 1.24977e-17 (rank : 24) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z1N6, Q91ZX9 | Gene names | Sfrp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related sequence protein 4 precursor (sFRP-4) (Frizzled-related protein 4) (FRP-4). | |||||
|
SFRP3_MOUSE
|
||||||
| NC score | 0.823411 (rank : 25) | θ value | 5.60996e-18 (rank : 23) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97401, O09075, O09093, Q91W58 | Gene names | Frzb, Fiz, Fre, Frzb1, Sfrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Secreted frizzled-related protein 3 precursor (sFRP-3) (Frizzled- related protein 1) (FrzB-1) (Frezzled) (Fritz). | |||||
|
SFRP3_HUMAN
|
||||||
| NC score | 0.822057 (rank : 26) | θ value | 3.63628e-17 (rank : 25) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92765, O00181, Q99686 | Gene names | FRZB, FIZ, FRE, FRP, FRZB1, SFRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Secreted frizzled-related protein 3 precursor (sFRP-3) (Frizzled- related protein 1) (FrzB-1) (Frezzled) (Fritz). | |||||
|
SFRP1_MOUSE
|
||||||
| NC score | 0.794290 (rank : 27) | θ value | 1.52774e-15 (rank : 27) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C4U3, O08861, Q8R1J4 | Gene names | Sfrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 1 precursor (sFRP-1). | |||||
|
SFRP1_HUMAN
|
||||||
| NC score | 0.792919 (rank : 28) | θ value | 3.40345e-15 (rank : 31) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N474, O00546, O14779 | Gene names | SFRP1, FRP, FRP1, SARP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 1 precursor (sFRP-1) (Frizzled- related protein 1) (FRP-1) (Secreted apoptosis-related protein 2) (SARP-2). | |||||
|
SFRP5_HUMAN
|
||||||
| NC score | 0.790280 (rank : 29) | θ value | 8.38298e-14 (rank : 33) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5T4F7, O14780, Q86TH7 | Gene names | SFRP5, FRP1B, SARP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5) (Secreted apoptosis-related protein 3) (SARP-3) (Frizzled-related protein 1b) (FRP-1b). | |||||
|
SFRP5_MOUSE
|
||||||
| NC score | 0.790067 (rank : 30) | θ value | 2.43908e-13 (rank : 34) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WU66, Q8K269 | Gene names | Sfrp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5). | |||||
|
SFRP2_MOUSE
|
||||||
| NC score | 0.789066 (rank : 31) | θ value | 2.60593e-15 (rank : 30) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97299, O08862, O35297 | Gene names | Sfrp2, Sarp1, Sdf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (SDF5 protein). | |||||
|
SFRP2_HUMAN
|
||||||
| NC score | 0.788177 (rank : 32) | θ value | 2.60593e-15 (rank : 29) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96HF1, O14778, Q9HAP5 | Gene names | SFRP2, FRP2, SARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (Frizzled-related protein 2) (FRP-2). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.441534 (rank : 33) | θ value | 3.18553e-13 (rank : 35) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.435046 (rank : 34) | θ value | 5.43371e-13 (rank : 36) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
CBPZ_HUMAN
|
||||||
| NC score | 0.418081 (rank : 35) | θ value | 3.64472e-09 (rank : 37) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.373576 (rank : 36) | θ value | 8.97725e-08 (rank : 38) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.278584 (rank : 37) | θ value | 2.60593e-15 (rank : 28) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CORIN_HUMAN
|
||||||
| NC score | 0.278168 (rank : 38) | θ value | 1.68911e-14 (rank : 32) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.243146 (rank : 39) | θ value | 0.00509761 (rank : 45) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GP157_HUMAN
|
||||||
| NC score | 0.186037 (rank : 40) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5UAW9, Q8WWB8, Q9HA73 | Gene names | GPR157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GP157_MOUSE
|
||||||
| NC score | 0.175916 (rank : 41) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C206, Q8C224 | Gene names | Gpr157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GP155_HUMAN
|
||||||
| NC score | 0.147519 (rank : 42) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.097864 (rank : 43) | θ value | 0.00869519 (rank : 46) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
BAI2_MOUSE
|
||||||
| NC score | 0.094257 (rank : 44) | θ value | 0.00298849 (rank : 42) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI2_HUMAN
|
||||||
| NC score | 0.094251 (rank : 45) | θ value | 0.00298849 (rank : 41) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.088477 (rank : 46) | θ value | 0.00175202 (rank : 40) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.085060 (rank : 47) | θ value | 0.000461057 (rank : 39) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI3_HUMAN
|
||||||
| NC score | 0.082809 (rank : 48) | θ value | 0.00509761 (rank : 43) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60242, O60297 | Gene names | BAI3, KIAA0550 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
BAI3_MOUSE
|
||||||
| NC score | 0.082624 (rank : 49) | θ value | 0.00509761 (rank : 44) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80ZF8 | Gene names | Bai3 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.066704 (rank : 50) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.061565 (rank : 51) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
LPHN3_HUMAN
|
||||||
| NC score | 0.060377 (rank : 52) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HAR2, O94867, Q9NWK5 | Gene names | LPHN3, KIAA0768, LEC3 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-3 precursor (Calcium-independent alpha-latrotoxin receptor 3) (Lectomedin-3). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.057120 (rank : 53) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GPR64_MOUSE
|
||||||
| NC score | 0.056120 (rank : 54) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.052265 (rank : 55) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.046849 (rank : 56) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
GP128_MOUSE
|
||||||
| NC score | 0.036667 (rank : 57) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BM96, Q80T42 | Gene names | Gpr128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
CECR6_HUMAN
|
||||||
| NC score | 0.036182 (rank : 58) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXQ6 | Gene names | CECR6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6. | |||||
|
CECR6_MOUSE
|
||||||
| NC score | 0.035643 (rank : 59) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MX7 | Gene names | Cecr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6 homolog. | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.035499 (rank : 60) | θ value | 0.0113563 (rank : 47) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.034370 (rank : 61) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.033085 (rank : 62) | θ value | 0.0252991 (rank : 49) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
SYNPR_MOUSE
|
||||||
| NC score | 0.031993 (rank : 63) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGN8, Q8C5J4, Q9DB89 | Gene names | Synpr | |||
|
Domain Architecture |

|
|||||
| Description | Synaptoporin. | |||||
|
SYNPR_HUMAN
|
||||||
| NC score | 0.031865 (rank : 64) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBG9 | Gene names | SYNPR | |||
|
Domain Architecture |

|
|||||
| Description | Synaptoporin. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.031328 (rank : 65) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.030931 (rank : 66) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.028360 (rank : 67) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CD97_HUMAN
|
||||||
| NC score | 0.026513 (rank : 68) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48960, O00718, O76101, Q8NG72, Q8TBQ7 | Gene names | CD97 | |||
|
Domain Architecture |

|
|||||
| Description | CD97 antigen precursor (Leukocyte antigen CD97). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.026231 (rank : 69) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.025783 (rank : 70) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
IG2AS_HUMAN
|
||||||
| NC score | 0.025580 (rank : 71) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6U949, Q7Z712, Q9P2W0 | Gene names | IGF2AS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative insulin-like growth factor 2 antisense gene protein (IGF2-AS) (PEG8/IGF2AS protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.024023 (rank : 72) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.022766 (rank : 73) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.022036 (rank : 74) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.021240 (rank : 75) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.020111 (rank : 76) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.020026 (rank : 77) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
S19A1_HUMAN
|
||||||
| NC score | 0.019825 (rank : 78) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41440, O00553, O60227, Q13026 | Gene names | SLC19A1, FLOT1, RFC1 | |||
|
Domain Architecture |

|
|||||
| Description | Folate transporter 1 (Solute carrier family 19 member 1) (Placental folate transporter) (FOLT) (Reduced folate carrier protein) (RFC) (Intestinal folate carrier) (IFC-1). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.019234 (rank : 79) | θ value | 0.0193708 (rank : 48) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.018951 (rank : 80) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.018546 (rank : 81) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.018492 (rank : 82) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.016778 (rank : 83) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.016185 (rank : 84) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.016124 (rank : 85) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
SYPL2_MOUSE
|
||||||
| NC score | 0.015883 (rank : 86) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89104 | Gene names | Sypl2, Mg29 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptophysin-like protein 2 (Mitsugumin-29) (Mg29). | |||||
|
SYPL2_HUMAN
|
||||||
| NC score | 0.015863 (rank : 87) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VXT5, Q6ZMX1 | Gene names | SYPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptophysin-like protein 2. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.015550 (rank : 88) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
REXO4_HUMAN
|
||||||
| NC score | 0.014148 (rank : 89) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR2, Q5T8S4, Q5T8S5, Q5T8S6, Q9GZW3 | Gene names | REXO4, PMC2, XPMC2H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (hPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.013720 (rank : 90) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.013720 (rank : 91) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.012607 (rank : 92) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.012215 (rank : 93) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.012123 (rank : 94) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.011437 (rank : 95) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.011381 (rank : 96) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
K0226_MOUSE
|
||||||
| NC score | 0.011268 (rank : 97) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.010855 (rank : 98) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BCAS1_HUMAN
|
||||||
| NC score | 0.009565 (rank : 99) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.008808 (rank : 100) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.007996 (rank : 101) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
PWWP2_HUMAN
|
||||||
| NC score | 0.007955 (rank : 102) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.007910 (rank : 103) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
RBM15_HUMAN
|
||||||
| NC score | 0.007869 (rank : 104) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.007700 (rank : 105) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.007415 (rank : 106) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.007094 (rank : 107) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.007042 (rank : 108) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.006874 (rank : 109) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.006126 (rank : 110) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
SFRS2_HUMAN
|
||||||
| NC score | 0.005944 (rank : 111) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| NC score | 0.005944 (rank : 112) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.005832 (rank : 113) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.005517 (rank : 114) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.005485 (rank : 115) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.005367 (rank : 116) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.005306 (rank : 117) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TLE3_MOUSE
|
||||||
| NC score | 0.005056 (rank : 118) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
IQEC3_HUMAN
|
||||||
| NC score | 0.004667 (rank : 119) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.004629 (rank : 120) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TLE3_HUMAN
|
||||||
| NC score | 0.004518 (rank : 121) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
TR136_MOUSE
|
||||||
| NC score | 0.004284 (rank : 122) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TQA8, Q7M716 | Gene names | Tas2r136, T2r52, Tas2r36 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 136 (T2R136) (Taste receptor type 2 member 36) (T2R36) (mT2r52). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.003533 (rank : 123) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.003312 (rank : 124) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.002977 (rank : 125) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.002311 (rank : 126) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
TLR5_HUMAN
|
||||||
| NC score | 0.001891 (rank : 127) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60602, O15456 | Gene names | TLR5, TIL3 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor (Toll/interleukin-1 receptor-like protein 3). | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.001871 (rank : 128) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
FIBA_HUMAN
|
||||||
| NC score | 0.001763 (rank : 129) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.001065 (rank : 130) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
PEG3_HUMAN
|
||||||
| NC score | -0.001743 (rank : 131) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
FOG1_HUMAN
|
||||||
| NC score | -0.002666 (rank : 132) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | -0.003190 (rank : 133) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
P2RY1_HUMAN
|
||||||
| NC score | -0.004024 (rank : 134) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47900 | Gene names | P2RY1 | |||
|
Domain Architecture |

|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
ZNF24_HUMAN
|
||||||
| NC score | -0.004668 (rank : 135) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17028, O14754 | Gene names | ZNF24, KOX17, ZNF191, ZSCAN3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 24 (Zinc finger protein 191) (Zinc finger protein KOX17) (Retinoic acid suppression protein A) (RSG-A) (Zinc finger and SCAN domain-containing protein 3). | |||||
|
ZN440_HUMAN
|
||||||
| NC score | -0.004708 (rank : 136) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYI8, Q8N1R9 | Gene names | ZNF440 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 440. | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | -0.004867 (rank : 137) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
M3K10_HUMAN
|
||||||
| NC score | -0.005210 (rank : 138) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||