Please be patient as the page loads
|
DNL3_MOUSE
|
||||||
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DNL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994632 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 2.04573e-44 (rank : 3) | NC score | 0.716456 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL1_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 4) | NC score | 0.739839 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL4_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 5) | NC score | 0.715810 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49917, Q8IY66, Q8TEU5 | Gene names | LIG4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
DNL4_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 6) | NC score | 0.707095 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTF7, Q3UG76 | Gene names | Lig4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 7) | NC score | 0.205467 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 8) | NC score | 0.201646 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.033468 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.033594 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
KLH20_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.015379 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2M5, Q5TZF2, Q9H457 | Gene names | KLHL20, KLEIP | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein) (Kelch- like protein X). | |||||
|
KLH20_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.015376 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCK5, Q8BWA2 | Gene names | Klhl20, Kleip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.036729 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.014133 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.014841 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.012531 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.021663 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.035835 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
TMCC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.023011 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.012077 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
TMCC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.022433 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZZ6, Q8CEF4 | Gene names | Tmcc1, Kiaa0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.045884 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.018675 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.013205 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.007736 (rank : 40) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.025729 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.010954 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
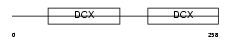
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
KLHL6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.012694 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ60, Q8N5I1, Q8N892 | Gene names | KLHL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 6. | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.025473 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.025280 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.025251 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.008428 (rank : 39) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.009955 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
KEAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.011523 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2X8 | Gene names | Keap1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like ECH-associated protein 1 (Cytosolic inhibitor of Nrf2). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.021140 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.010837 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
NTBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.013452 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7V8, Q49A22, Q6P1P9, Q9HAE6, Q9Y350 | Gene names | SCYL1BP1, NTKLBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (hNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
PARP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.069124 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.069153 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.067042 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
DNL3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.994632 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL1_MOUSE
|
||||||
| NC score | 0.739839 (rank : 3) | θ value | 2.50074e-42 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.716456 (rank : 4) | θ value | 2.04573e-44 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DNL4_HUMAN
|
||||||
| NC score | 0.715810 (rank : 5) | θ value | 6.59618e-35 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49917, Q8IY66, Q8TEU5 | Gene names | LIG4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
DNL4_MOUSE
|
||||||
| NC score | 0.707095 (rank : 6) | θ value | 1.52067e-31 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTF7, Q3UG76 | Gene names | Lig4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA ligase 4 (EC 6.5.1.1) (DNA ligase IV) (Polydeoxyribonucleotide synthase [ATP] 4). | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.205467 (rank : 7) | θ value | 7.59969e-07 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP1_HUMAN
|
||||||
| NC score | 0.201646 (rank : 8) | θ value | 1.29631e-06 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP2_MOUSE
|
||||||
| NC score | 0.069153 (rank : 9) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP2_HUMAN
|
||||||
| NC score | 0.069124 (rank : 10) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| NC score | 0.067042 (rank : 11) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.045884 (rank : 12) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.036729 (rank : 13) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.035835 (rank : 14) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.033594 (rank : 15) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.033468 (rank : 16) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.025729 (rank : 17) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.025473 (rank : 18) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.025280 (rank : 19) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.025251 (rank : 20) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
TMCC1_HUMAN
|
||||||
| NC score | 0.023011 (rank : 21) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94876, Q68E06, Q8IXM8 | Gene names | TMCC1, KIAA0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
TMCC1_MOUSE
|
||||||
| NC score | 0.022433 (rank : 22) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZZ6, Q8CEF4 | Gene names | Tmcc1, Kiaa0779 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 1. | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.021663 (rank : 23) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.021140 (rank : 24) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.018675 (rank : 25) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
KLH20_HUMAN
|
||||||
| NC score | 0.015379 (rank : 26) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2M5, Q5TZF2, Q9H457 | Gene names | KLHL20, KLEIP | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein) (Kelch- like protein X). | |||||
|
KLH20_MOUSE
|
||||||
| NC score | 0.015376 (rank : 27) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCK5, Q8BWA2 | Gene names | Klhl20, Kleip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.014841 (rank : 28) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.014133 (rank : 29) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
NTBP1_HUMAN
|
||||||
| NC score | 0.013452 (rank : 30) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7V8, Q49A22, Q6P1P9, Q9HAE6, Q9Y350 | Gene names | SCYL1BP1, NTKLBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (hNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.013205 (rank : 31) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
KLHL6_HUMAN
|
||||||
| NC score | 0.012694 (rank : 32) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ60, Q8N5I1, Q8N892 | Gene names | KLHL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 6. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.012531 (rank : 33) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.012077 (rank : 34) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
KEAP1_MOUSE
|
||||||
| NC score | 0.011523 (rank : 35) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2X8 | Gene names | Keap1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like ECH-associated protein 1 (Cytosolic inhibitor of Nrf2). | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.010954 (rank : 36) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.010837 (rank : 37) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.009955 (rank : 38) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.008428 (rank : 39) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.007736 (rank : 40) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||