Please be patient as the page loads
|
KCTD8_HUMAN
|
||||||
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCTD8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCTD8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996288 (rank : 2) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCD16_HUMAN
|
||||||
| θ value | 9.56396e-135 (rank : 3) | NC score | 0.983692 (rank : 4) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q68DU8, Q9P2M9 | Gene names | KCTD16, KIAA1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD16_MOUSE
|
||||||
| θ value | 3.07662e-133 (rank : 4) | NC score | 0.984109 (rank : 3) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5DTY9, Q78Y50 | Gene names | Kctd16, Kiaa1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD12_HUMAN
|
||||||
| θ value | 2.31364e-80 (rank : 5) | NC score | 0.968267 (rank : 5) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD12_MOUSE
|
||||||
| θ value | 1.14825e-79 (rank : 6) | NC score | 0.967605 (rank : 6) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCTD4_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 7) | NC score | 0.746901 (rank : 12) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 8) | NC score | 0.746665 (rank : 13) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD6_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 9) | NC score | 0.749583 (rank : 10) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 10) | NC score | 0.749462 (rank : 11) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCNRG_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 11) | NC score | 0.766263 (rank : 8) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q2TUM3, Q6P8J6, Q8C6V1 | Gene names | Kcnrg, Clld4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCNRG_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 12) | NC score | 0.763015 (rank : 9) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N5I3, Q8IU75, Q8N3Q9 | Gene names | KCNRG, CLLD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCTD7_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 13) | NC score | 0.693685 (rank : 15) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCTD7_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 14) | NC score | 0.695053 (rank : 14) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCD14_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 15) | NC score | 0.766422 (rank : 7) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BQ13 | Gene names | KCTD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD14. | |||||
|
KCD15_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 16) | NC score | 0.691175 (rank : 16) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCD15_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 17) | NC score | 0.690344 (rank : 17) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD1_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 18) | NC score | 0.677753 (rank : 19) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 19) | NC score | 0.677753 (rank : 20) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCD10_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 20) | NC score | 0.585459 (rank : 25) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H3F6, Q53HN2, Q59FV1, Q6PL47, Q96SU0 | Gene names | KCTD10, ULR061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCD18_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 21) | NC score | 0.690269 (rank : 18) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KCD10_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 22) | NC score | 0.577891 (rank : 27) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q922M3 | Gene names | Kctd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCTD5_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 23) | NC score | 0.595488 (rank : 21) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NXV2 | Gene names | KCTD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD5_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 24) | NC score | 0.594069 (rank : 22) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VC57 | Gene names | Kctd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD3_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 25) | NC score | 0.565136 (rank : 30) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y597, Q49AG7, Q504Q9, Q6PJN6, Q8ND58, Q8NDJ0, Q8WX16 | Gene names | KCTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3 (NY-REN-45 antigen). | |||||
|
KCD13_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 26) | NC score | 0.564929 (rank : 31) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ19, Q96P93, Q96SA1 | Gene names | KCTD13, PDIP1, POLDIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1) (TNFAIP1-like protein). | |||||
|
KCTD3_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 27) | NC score | 0.561625 (rank : 35) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
TNAP1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 28) | NC score | 0.564715 (rank : 33) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13829 | Gene names | TNFAIP1, EDP1 | |||
|
Domain Architecture |
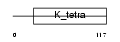
|
|||||
| Description | BTB/POZ domain-containing protein TNFAIP1 (Tumor necrosis factor, alpha-induced protein 1, endothelial) (Protein B12). | |||||
|
KCD13_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 29) | NC score | 0.562185 (rank : 34) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGV7, Q6AXE9 | Gene names | Kctd13, Poldip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1). | |||||
|
KCTD2_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 30) | NC score | 0.573347 (rank : 28) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14681 | Gene names | KCTD2, KIAA0176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCTD9_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 31) | NC score | 0.585104 (rank : 26) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCTD9_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 32) | NC score | 0.593313 (rank : 23) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCD17_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 33) | NC score | 0.585753 (rank : 24) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
KCD11_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 34) | NC score | 0.571675 (rank : 29) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q693B1 | Gene names | KCTD11, REN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCD11_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 35) | NC score | 0.564803 (rank : 32) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K485 | Gene names | Kctd11, Ren | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCTD2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 36) | NC score | 0.465589 (rank : 36) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CEZ0 | Gene names | Kctd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 37) | NC score | 0.179665 (rank : 43) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 38) | NC score | 0.174418 (rank : 44) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 39) | NC score | 0.187239 (rank : 41) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 40) | NC score | 0.187183 (rank : 42) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 41) | NC score | 0.162008 (rank : 47) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 42) | NC score | 0.162078 (rank : 46) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 43) | NC score | 0.190342 (rank : 40) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 44) | NC score | 0.190380 (rank : 39) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.060936 (rank : 76) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 46) | NC score | 0.060936 (rank : 77) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 47) | NC score | 0.162845 (rank : 45) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.201121 (rank : 37) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.136456 (rank : 48) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.054543 (rank : 78) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.198395 (rank : 38) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.017950 (rank : 85) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.028573 (rank : 80) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.007334 (rank : 102) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.014481 (rank : 89) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
MERTK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.003379 (rank : 111) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12866, Q9HBB4 | Gene names | MERTK, MER | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
SIP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.003571 (rank : 110) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60315, Q9UED1 | Gene names | ZFHX1B, KIAA0569, SIP1, ZFX1B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1) (SMADIP1). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.019196 (rank : 83) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.030278 (rank : 79) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.118278 (rank : 49) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.008122 (rank : 99) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
MERTK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.001380 (rank : 115) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60805, Q62194 | Gene names | Mertk, Mer | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.005938 (rank : 106) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.002785 (rank : 113) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.013192 (rank : 90) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.009469 (rank : 96) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
2A5D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.010811 (rank : 92) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14738, O00494, O00696, Q15171 | Gene names | PPP2R5D | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (PP2A, B subunit, B' delta isoform) (PP2A, B subunit, B56 delta isoform) (PP2A, B subunit, PR61 delta isoform) (PP2A, B subunit, R5 delta isoform). | |||||
|
ES8L3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.018334 (rank : 84) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WL0, Q3TJ14 | Gene names | Eps8l3, Eps8r3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor pathway substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.008699 (rank : 98) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TISD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.019471 (rank : 82) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
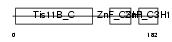
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TLE4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.003229 (rank : 112) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.017090 (rank : 86) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.010038 (rank : 94) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CXA8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.006408 (rank : 104) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48165, Q9NP25 | Gene names | GJA8 | |||
|
Domain Architecture |
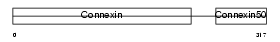
|
|||||
| Description | Gap junction alpha-8 protein (Connexin-50) (Cx50) (Lens fiber protein MP70). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.006139 (rank : 105) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.005925 (rank : 107) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.005044 (rank : 108) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RT32_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.017090 (rank : 87) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPV3 | Gene names | Mrps32, D10Ertd322e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S32 (S32mt) (MRP-S32). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.009461 (rank : 97) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
ALX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | -0.000023 (rank : 116) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H161, Q96JN7, Q9H198, Q9HAY9 | Gene names | ALX4, KIAA1788 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.014763 (rank : 88) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.009944 (rank : 95) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.010850 (rank : 91) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SI1L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.007998 (rank : 100) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.001540 (rank : 114) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.020588 (rank : 81) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.010111 (rank : 93) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.006429 (rank : 103) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.115775 (rank : 50) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.004211 (rank : 109) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.007784 (rank : 101) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.080802 (rank : 67) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.081202 (rank : 66) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.080765 (rank : 68) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.080222 (rank : 72) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.080753 (rank : 69) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.080257 (rank : 71) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.088984 (rank : 57) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.088894 (rank : 58) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.081747 (rank : 64) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.081242 (rank : 65) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.084672 (rank : 61) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.084676 (rank : 60) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.092430 (rank : 55) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.093674 (rank : 54) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.088770 (rank : 59) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.113178 (rank : 51) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.066339 (rank : 75) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.092108 (rank : 56) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.069865 (rank : 74) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.083081 (rank : 62) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.082750 (rank : 63) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.080593 (rank : 70) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.072727 (rank : 73) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.099575 (rank : 52) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.098832 (rank : 53) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCTD8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCTD8_MOUSE
|
||||||
| NC score | 0.996288 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCD16_MOUSE
|
||||||
| NC score | 0.984109 (rank : 3) | θ value | 3.07662e-133 (rank : 4) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5DTY9, Q78Y50 | Gene names | Kctd16, Kiaa1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD16_HUMAN
|
||||||
| NC score | 0.983692 (rank : 4) | θ value | 9.56396e-135 (rank : 3) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q68DU8, Q9P2M9 | Gene names | KCTD16, KIAA1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD12_HUMAN
|
||||||
| NC score | 0.968267 (rank : 5) | θ value | 2.31364e-80 (rank : 5) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD12_MOUSE
|
||||||
| NC score | 0.967605 (rank : 6) | θ value | 1.14825e-79 (rank : 6) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD14_HUMAN
|
||||||
| NC score | 0.766422 (rank : 7) | θ value | 5.43371e-13 (rank : 15) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BQ13 | Gene names | KCTD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD14. | |||||
|
KCNRG_MOUSE
|
||||||
| NC score | 0.766263 (rank : 8) | θ value | 6.41864e-14 (rank : 11) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q2TUM3, Q6P8J6, Q8C6V1 | Gene names | Kcnrg, Clld4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCNRG_HUMAN
|
||||||
| NC score | 0.763015 (rank : 9) | θ value | 1.09485e-13 (rank : 12) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N5I3, Q8IU75, Q8N3Q9 | Gene names | KCNRG, CLLD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCTD6_HUMAN
|
||||||
| NC score | 0.749583 (rank : 10) | θ value | 9.90251e-15 (rank : 9) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_MOUSE
|
||||||
| NC score | 0.749462 (rank : 11) | θ value | 9.90251e-15 (rank : 10) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD4_HUMAN
|
||||||
| NC score | 0.746901 (rank : 12) | θ value | 7.58209e-15 (rank : 7) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_MOUSE
|
||||||
| NC score | 0.746665 (rank : 13) | θ value | 7.58209e-15 (rank : 8) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD7_HUMAN
|
||||||
| NC score | 0.695053 (rank : 14) | θ value | 1.86753e-13 (rank : 14) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCTD7_MOUSE
|
||||||
| NC score | 0.693685 (rank : 15) | θ value | 1.42992e-13 (rank : 13) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCD15_MOUSE
|
||||||
| NC score | 0.691175 (rank : 16) | θ value | 7.09661e-13 (rank : 16) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCD15_HUMAN
|
||||||
| NC score | 0.690344 (rank : 17) | θ value | 1.58096e-12 (rank : 17) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCD18_HUMAN
|
||||||
| NC score | 0.690269 (rank : 18) | θ value | 6.21693e-09 (rank : 21) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KCTD1_HUMAN
|
||||||
| NC score | 0.677753 (rank : 19) | θ value | 1.02475e-11 (rank : 18) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| NC score | 0.677753 (rank : 20) | θ value | 1.02475e-11 (rank : 19) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD5_HUMAN
|
||||||
| NC score | 0.595488 (rank : 21) | θ value | 4.0297e-08 (rank : 23) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NXV2 | Gene names | KCTD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD5_MOUSE
|
||||||
| NC score | 0.594069 (rank : 22) | θ value | 5.26297e-08 (rank : 24) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VC57 | Gene names | Kctd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD9_MOUSE
|
||||||
| NC score | 0.593313 (rank : 23) | θ value | 6.43352e-06 (rank : 32) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCD17_HUMAN
|
||||||
| NC score | 0.585753 (rank : 24) | θ value | 1.09739e-05 (rank : 33) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
KCD10_HUMAN
|
||||||
| NC score | 0.585459 (rank : 25) | θ value | 2.79066e-09 (rank : 20) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H3F6, Q53HN2, Q59FV1, Q6PL47, Q96SU0 | Gene names | KCTD10, ULR061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCTD9_HUMAN
|
||||||
| NC score | 0.585104 (rank : 26) | θ value | 6.43352e-06 (rank : 31) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCD10_MOUSE
|
||||||
| NC score | 0.577891 (rank : 27) | θ value | 8.11959e-09 (rank : 22) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q922M3 | Gene names | Kctd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCTD2_HUMAN
|
||||||
| NC score | 0.573347 (rank : 28) | θ value | 5.81887e-07 (rank : 30) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14681 | Gene names | KCTD2, KIAA0176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCD11_HUMAN
|
||||||
| NC score | 0.571675 (rank : 29) | θ value | 0.000158464 (rank : 34) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q693B1 | Gene names | KCTD11, REN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCTD3_HUMAN
|
||||||
| NC score | 0.565136 (rank : 30) | θ value | 8.97725e-08 (rank : 25) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y597, Q49AG7, Q504Q9, Q6PJN6, Q8ND58, Q8NDJ0, Q8WX16 | Gene names | KCTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3 (NY-REN-45 antigen). | |||||
|
KCD13_HUMAN
|
||||||
| NC score | 0.564929 (rank : 31) | θ value | 1.53129e-07 (rank : 26) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ19, Q96P93, Q96SA1 | Gene names | KCTD13, PDIP1, POLDIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1) (TNFAIP1-like protein). | |||||
|
KCD11_MOUSE
|
||||||
| NC score | 0.564803 (rank : 32) | θ value | 0.00035302 (rank : 35) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K485 | Gene names | Kctd11, Ren | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
TNAP1_HUMAN
|
||||||
| NC score | 0.564715 (rank : 33) | θ value | 3.41135e-07 (rank : 28) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13829 | Gene names | TNFAIP1, EDP1 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein TNFAIP1 (Tumor necrosis factor, alpha-induced protein 1, endothelial) (Protein B12). | |||||
|
KCD13_MOUSE
|
||||||
| NC score | 0.562185 (rank : 34) | θ value | 5.81887e-07 (rank : 29) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGV7, Q6AXE9 | Gene names | Kctd13, Poldip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1). | |||||
|
KCTD3_MOUSE
|
||||||
| NC score | 0.561625 (rank : 35) | θ value | 3.41135e-07 (rank : 27) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
KCTD2_MOUSE
|
||||||
| NC score | 0.465589 (rank : 36) | θ value | 0.00134147 (rank : 36) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CEZ0 | Gene names | Kctd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.201121 (rank : 37) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.198395 (rank : 38) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.190380 (rank : 39) | θ value | 0.0252991 (rank : 44) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.190342 (rank : 40) | θ value | 0.0252991 (rank : 43) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.187239 (rank : 41) | θ value | 0.00869519 (rank : 39) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.187183 (rank : 42) | θ value | 0.00869519 (rank : 40) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.179665 (rank : 43) | θ value | 0.00228821 (rank : 37) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.174418 (rank : 44) | θ value | 0.00869519 (rank : 38) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.162845 (rank : 45) | θ value | 0.0961366 (rank : 47) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.162078 (rank : 46) | θ value | 0.0148317 (rank : 42) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.162008 (rank : 47) | θ value | 0.0148317 (rank : 41) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.136456 (rank : 48) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.118278 (rank : 49) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.115775 (rank : 50) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.113178 (rank : 51) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.099575 (rank : 52) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.098832 (rank : 53) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.093674 (rank : 54) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.092430 (rank : 55) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.092108 (rank : 56) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.088984 (rank : 57) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.088894 (rank : 58) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.088770 (rank : 59) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.084676 (rank : 60) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.084672 (rank : 61) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.083081 (rank : 62) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.082750 (rank : 63) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.081747 (rank : 64) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.081242 (rank : 65) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.081202 (rank : 66) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.080802 (rank : 67) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.080765 (rank : 68) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.080753 (rank : 69) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.080593 (rank : 70) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.080257 (rank : 71) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.080222 (rank : 72) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.072727 (rank : 73) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.069865 (rank : 74) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.066339 (rank : 75) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.060936 (rank : 76) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.060936 (rank : 77) | θ value | 0.0736092 (rank : 46) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.054543 (rank : 78) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.030278 (rank : 79) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.028573 (rank : 80) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.020588 (rank : 81) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.019471 (rank : 82) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.019196 (rank : 83) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ES8L3_MOUSE
|
||||||
| NC score | 0.018334 (rank : 84) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WL0, Q3TJ14 | Gene names | Eps8l3, Eps8r3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor pathway substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.017950 (rank : 85) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.017090 (rank : 86) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
RT32_MOUSE
|
||||||
| NC score | 0.017090 (rank : 87) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPV3 | Gene names | Mrps32, D10Ertd322e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S32 (S32mt) (MRP-S32). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.014763 (rank : 88) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.014481 (rank : 89) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.013192 (rank : 90) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.010850 (rank : 91) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
2A5D_HUMAN
|
||||||
| NC score | 0.010811 (rank : 92) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14738, O00494, O00696, Q15171 | Gene names | PPP2R5D | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform (PP2A, B subunit, B' delta isoform) (PP2A, B subunit, B56 delta isoform) (PP2A, B subunit, PR61 delta isoform) (PP2A, B subunit, R5 delta isoform). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.010111 (rank : 93) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.010038 (rank : 94) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.009944 (rank : 95) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.009469 (rank : 96) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.009461 (rank : 97) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.008699 (rank : 98) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.008122 (rank : 99) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
SI1L2_MOUSE
|
||||||
| NC score | 0.007998 (rank : 100) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.007784 (rank : 101) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.007334 (rank : 102) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.006429 (rank : 103) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
CXA8_HUMAN
|
||||||
| NC score | 0.006408 (rank : 104) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48165, Q9NP25 | Gene names | GJA8 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-8 protein (Connexin-50) (Cx50) (Lens fiber protein MP70). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.006139 (rank : 105) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.005938 (rank : 106) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.005925 (rank : 107) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.005044 (rank : 108) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.004211 (rank : 109) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
SIP1_HUMAN
|
||||||
| NC score | 0.003571 (rank : 110) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60315, Q9UED1 | Gene names | ZFHX1B, KIAA0569, SIP1, ZFX1B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1) (SMADIP1). | |||||
|
MERTK_HUMAN
|
||||||
| NC score | 0.003379 (rank : 111) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12866, Q9HBB4 | Gene names | MERTK, MER | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
TLE4_MOUSE
|
||||||
| NC score | 0.003229 (rank : 112) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.002785 (rank : 113) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
SIP1_MOUSE
|
||||||
| NC score | 0.001540 (rank : 114) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
MERTK_MOUSE
|
||||||
| NC score | 0.001380 (rank : 115) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60805, Q62194 | Gene names | Mertk, Mer | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
ALX4_HUMAN
|
||||||
| NC score | -0.000023 (rank : 116) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 91 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H161, Q96JN7, Q9H198, Q9HAY9 | Gene names | ALX4, KIAA1788 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4. | |||||