Please be patient as the page loads
|
SI1L2_MOUSE
|
||||||
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SI1L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.978058 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975455 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996277 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.957882 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 1.01004e-176 (rank : 6) | NC score | 0.933242 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 6.77497e-173 (rank : 7) | NC score | 0.942993 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 2.32979e-56 (rank : 8) | NC score | 0.885562 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 9) | NC score | 0.462192 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 10) | NC score | 0.449222 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 11) | NC score | 0.436518 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 12) | NC score | 0.415387 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 13) | NC score | 0.101717 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.131557 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
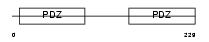
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.125547 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.141617 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.141584 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.126227 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.061024 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.144613 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.083799 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.131793 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.125323 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.111295 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.027224 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.036696 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.053255 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.107404 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.104821 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.063387 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.038832 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
APBA3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.138737 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |
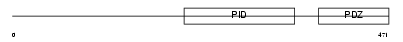
|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.102224 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.058346 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.037138 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.036974 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.049539 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.032999 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.094888 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.032659 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.003185 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.025723 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.025276 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.037701 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.048026 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.031518 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.031128 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.031744 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.020426 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
REST_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.026978 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.027793 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.011330 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LYN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.002937 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07948 | Gene names | LYN | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Lyn (EC 2.7.10.2). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.034017 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.079984 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
RN103_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.022739 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00237, Q8IVB9 | Gene names | RNF103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 103 (Zinc finger protein 103 homolog) (Zfp-103) (KF-1) (hKF-1). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.037875 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.044510 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.033764 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CO029_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.021118 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H079, Q2TAC0, Q9H670 | Gene names | C15orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf29. | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.015149 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.061600 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.058690 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.056884 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.032253 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
DMPK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.003566 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
FCRL5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.010639 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RD9, Q495Q2, Q495Q4, Q5VYK9, Q6UY46 | Gene names | FCRL5, FCRH5, IRTA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 5 precursor (Immunoglobulin receptor translocation-associated gene 2 protein) (BXMAS1) (CD307 antigen). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.034898 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
K1967_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.032664 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VDP4, Q6PIB1, Q8BWR5, Q8C0F0, Q8R3G6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1967 homolog. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.033463 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PARVB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.011902 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBI1, Q5TGJ5, Q86X93, Q9NSP7, Q9UGT3, Q9Y368, Q9Y3L6, Q9Y3L7 | Gene names | PARVB | |||
|
Domain Architecture |
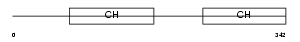
|
|||||
| Description | Beta-parvin (Affixin). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.086869 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.034467 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.044187 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.022695 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.029041 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.033986 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.019017 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.044267 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.017382 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.026658 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
LYN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.001328 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25911, Q62127 | Gene names | Lyn | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Lyn (EC 2.7.10.2). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.054780 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.064159 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.062044 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.079013 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.000456 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TTLL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.009442 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |
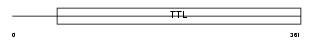
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.026626 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.028768 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.014674 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
LMNB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.022558 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.010457 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.030277 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.035064 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.023332 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.033569 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.074623 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.072551 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.011808 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.021448 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.046237 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.029306 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
FES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | -0.000336 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
KCTD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.007998 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.018795 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.024362 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
SDCB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.046502 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |
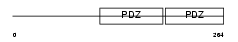
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.025018 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.025240 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.001516 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.005185 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.028685 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.023868 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DHB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.003827 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51658, O08898 | Gene names | Hsd17b2, Edh17b2 | |||
|
Domain Architecture |
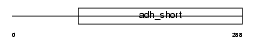
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (17- beta-hydroxysteroid dehydrogenase 2). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.001606 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.020343 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
LMNB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.020644 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.018032 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.007939 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.071803 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.041183 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.003266 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.021610 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.069655 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053872 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
SDCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.051626 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H190, O95892, Q5W0X1, Q9BZ42, Q9H567, Q9NRY8 | Gene names | SDCBP2, SITAC18 | |||
|
Domain Architecture |
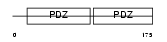
|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
SI1L2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.996277 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.978058 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.975455 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.957882 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.942993 (rank : 6) | θ value | 6.77497e-173 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.933242 (rank : 7) | θ value | 1.01004e-176 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.885562 (rank : 8) | θ value | 2.32979e-56 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| NC score | 0.462192 (rank : 9) | θ value | 4.16044e-13 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| NC score | 0.449222 (rank : 10) | θ value | 3.52202e-12 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.436518 (rank : 11) | θ value | 1.47974e-10 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.415387 (rank : 12) | θ value | 3.64472e-09 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.144613 (rank : 13) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.141617 (rank : 14) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.141584 (rank : 15) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA3_MOUSE
|
||||||
| NC score | 0.138737 (rank : 16) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.131793 (rank : 17) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.131557 (rank : 18) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.126227 (rank : 19) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.125547 (rank : 20) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.125323 (rank : 21) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.111295 (rank : 22) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.107404 (rank : 23) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.104821 (rank : 24) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.102224 (rank : 25) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.101717 (rank : 26) | θ value | 0.000270298 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.094888 (rank : 27) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.086869 (rank : 28) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.083799 (rank : 29) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.079984 (rank : 30) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.079013 (rank : 31) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.074623 (rank : 32) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.072551 (rank : 33) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.071803 (rank : 34) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.069655 (rank : 35) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.064159 (rank : 36) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.063387 (rank : 37) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.062044 (rank : 38) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.061600 (rank : 39) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.061024 (rank : 40) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.058690 (rank : 41) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.058346 (rank : 42) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.056884 (rank : 43) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.054780 (rank : 44) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
PDZD7_HUMAN
|
||||||
| NC score | 0.053872 (rank : 45) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.053255 (rank : 46) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
SDCB2_HUMAN
|
||||||
| NC score | 0.051626 (rank : 47) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H190, O95892, Q5W0X1, Q9BZ42, Q9H567, Q9NRY8 | Gene names | SDCBP2, SITAC18 | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.049539 (rank : 48) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.048026 (rank : 49) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
SDCB1_HUMAN
|
||||||
| NC score | 0.046502 (rank : 50) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.046237 (rank : 51) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.044510 (rank : 52) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.044267 (rank : 53) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.044187 (rank : 54) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.041183 (rank : 55) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.038832 (rank : 56) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.037875 (rank : 57) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.037701 (rank : 58) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.037138 (rank : 59) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.036974 (rank : 60) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.036696 (rank : 61) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.035064 (rank : 62) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.034898 (rank : 63) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.034467 (rank : 64) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.034017 (rank : 65) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.033986 (rank : 66) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.033764 (rank : 67) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.033569 (rank : 68) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.033463 (rank : 69) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.032999 (rank : 70) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
K1967_MOUSE
|
||||||
| NC score | 0.032664 (rank : 71) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VDP4, Q6PIB1, Q8BWR5, Q8C0F0, Q8R3G6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1967 homolog. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.032659 (rank : 72) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.032253 (rank : 73) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.031744 (rank : 74) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.031518 (rank : 75) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.031128 (rank : 76) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.030277 (rank : 77) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.029306 (rank : 78) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.029041 (rank : 79) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.028768 (rank : 80) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.028685 (rank : 81) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.027793 (rank : 82) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.027224 (rank : 83) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.026978 (rank : 84) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.026658 (rank : 85) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.026626 (rank : 86) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.025723 (rank : 87) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.025276 (rank : 88) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.025240 (rank : 89) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.025018 (rank : 90) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.024362 (rank : 91) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.023868 (rank : 92) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.023332 (rank : 93) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
RN103_HUMAN
|
||||||
| NC score | 0.022739 (rank : 94) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00237, Q8IVB9 | Gene names | RNF103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 103 (Zinc finger protein 103 homolog) (Zfp-103) (KF-1) (hKF-1). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.022695 (rank : 95) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
LMNB2_MOUSE
|
||||||
| NC score | 0.022558 (rank : 96) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.021610 (rank : 97) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.021448 (rank : 98) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CO029_HUMAN
|
||||||
| NC score | 0.021118 (rank : 99) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H079, Q2TAC0, Q9H670 | Gene names | C15orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf29. | |||||
|
LMNB2_HUMAN
|
||||||
| NC score | 0.020644 (rank : 100) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.020426 (rank : 101) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.020343 (rank : 102) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.019017 (rank : 103) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.018795 (rank : 104) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.018032 (rank : 105) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.017382 (rank : 106) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.015149 (rank : 107) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.014674 (rank : 108) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PARVB_HUMAN
|
||||||
| NC score | 0.011902 (rank : 109) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBI1, Q5TGJ5, Q86X93, Q9NSP7, Q9UGT3, Q9Y368, Q9Y3L6, Q9Y3L7 | Gene names | PARVB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-parvin (Affixin). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.011808 (rank : 110) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.011330 (rank : 111) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
FCRL5_HUMAN
|
||||||
| NC score | 0.010639 (rank : 112) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RD9, Q495Q2, Q495Q4, Q5VYK9, Q6UY46 | Gene names | FCRL5, FCRH5, IRTA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 5 precursor (Immunoglobulin receptor translocation-associated gene 2 protein) (BXMAS1) (CD307 antigen). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.010457 (rank : 113) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
TTLL1_MOUSE
|
||||||
| NC score | 0.009442 (rank : 114) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
KCTD8_HUMAN
|
||||||
| NC score | 0.007998 (rank : 115) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.007939 (rank : 116) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.005185 (rank : 117) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
DHB2_MOUSE
|
||||||
| NC score | 0.003827 (rank : 118) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51658, O08898 | Gene names | Hsd17b2, Edh17b2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (17- beta-hydroxysteroid dehydrogenase 2). | |||||
|
DMPK_HUMAN
|
||||||
| NC score | 0.003566 (rank : 119) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.003266 (rank : 120) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.003185 (rank : 121) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
LYN_HUMAN
|
||||||
| NC score | 0.002937 (rank : 122) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07948 | Gene names | LYN | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Lyn (EC 2.7.10.2). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.001606 (rank : 123) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.001516 (rank : 124) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
LYN_MOUSE
|
||||||
| NC score | 0.001328 (rank : 125) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25911, Q62127 | Gene names | Lyn | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Lyn (EC 2.7.10.2). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.000456 (rank : 126) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
FES_HUMAN
|
||||||
| NC score | -0.000336 (rank : 127) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||