Please be patient as the page loads
|
SI1L1_HUMAN
|
||||||
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SI1L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 162 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994143 (rank : 2) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.977285 (rank : 4) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.978058 (rank : 3) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.954631 (rank : 5) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 6.75929e-181 (rank : 6) | NC score | 0.929786 (rank : 7) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 9.45368e-175 (rank : 7) | NC score | 0.940008 (rank : 6) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 9.45883e-58 (rank : 8) | NC score | 0.883970 (rank : 8) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 9) | NC score | 0.458304 (rank : 9) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 10) | NC score | 0.445664 (rank : 10) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 11) | NC score | 0.432395 (rank : 11) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 12) | NC score | 0.410810 (rank : 12) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 13) | NC score | 0.101111 (rank : 24) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.055499 (rank : 41) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.145236 (rank : 13) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 16) | NC score | 0.131771 (rank : 17) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.089154 (rank : 27) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.136433 (rank : 15) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.136462 (rank : 14) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.041946 (rank : 59) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
PARD3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.082732 (rank : 29) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.047936 (rank : 50) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.121276 (rank : 18) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.038430 (rank : 69) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
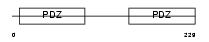
NHERF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.120604 (rank : 19) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
APBA3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.134827 (rank : 16) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |
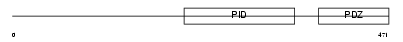
|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.072528 (rank : 31) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.019915 (rank : 121) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.113470 (rank : 21) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.100948 (rank : 25) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.120032 (rank : 20) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.036294 (rank : 74) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.035223 (rank : 76) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.101843 (rank : 22) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.101542 (rank : 23) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.053452 (rank : 44) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
VAPB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.052145 (rank : 46) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QY76 | Gene names | Vapb | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B (VAMP- associated protein B) (VAMP-associated protein 33b) (VAMP-B) (VAP-B). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.042918 (rank : 56) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.042410 (rank : 58) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.046346 (rank : 52) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.030362 (rank : 92) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.030276 (rank : 93) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.044151 (rank : 55) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.040445 (rank : 63) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
VAPB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.061214 (rank : 36) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95292, O95293, Q9P0H0 | Gene names | VAPB | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B/C (VAMP- associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.058454 (rank : 38) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.039427 (rank : 65) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.039018 (rank : 67) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.036615 (rank : 73) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.032994 (rank : 80) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.084870 (rank : 28) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.056305 (rank : 40) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.020094 (rank : 120) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.060236 (rank : 37) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.057977 (rank : 39) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.042466 (rank : 57) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.020221 (rank : 119) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.026344 (rank : 108) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.032675 (rank : 81) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.016925 (rank : 129) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
VAPA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.041747 (rank : 60) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P0L0, O75453, Q5U0E7, Q9UBZ2 | Gene names | VAPA, VAP33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
VAPA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.041486 (rank : 61) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV55, Q3TJM1, Q9QY77 | Gene names | Vapa, Vap33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.090692 (rank : 26) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
NSDHL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.023050 (rank : 114) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1J0, O55109 | Gene names | Nsdhl | |||
|
Domain Architecture |

|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170). | |||||
|
SBDS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.029925 (rank : 95) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3A5, Q96FX0, Q9NV53 | Gene names | SBDS | |||
|
Domain Architecture |

|
|||||
| Description | Shwachman-Bodian-Diamond syndrome protein. | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.010868 (rank : 142) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.068903 (rank : 33) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.029302 (rank : 101) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.047022 (rank : 51) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.021830 (rank : 115) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.029562 (rank : 99) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.055218 (rank : 42) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.045559 (rank : 54) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.037667 (rank : 70) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.014407 (rank : 133) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.040248 (rank : 64) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.033706 (rank : 79) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.033882 (rank : 78) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
MACOI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.036252 (rank : 75) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.052545 (rank : 45) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.050523 (rank : 49) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
SBDS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.027233 (rank : 106) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70122, Q3TG78, Q9CR18 | Gene names | Sbds | |||
|
Domain Architecture |
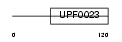
|
|||||
| Description | Shwachman-Bodian-Diamond syndrome protein homolog (Protein 22A3). | |||||
|
THAP6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.046031 (rank : 53) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |
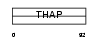
|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.039357 (rank : 66) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.024271 (rank : 111) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.034019 (rank : 77) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.037353 (rank : 71) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.036827 (rank : 72) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.009828 (rank : 143) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.040987 (rank : 62) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.028531 (rank : 103) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.029584 (rank : 98) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.021060 (rank : 118) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.001191 (rank : 161) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
OTOF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.012035 (rank : 135) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.078498 (rank : 30) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
ST5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.007305 (rank : 149) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.070884 (rank : 32) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.029686 (rank : 97) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
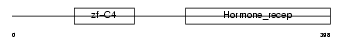
COT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.004868 (rank : 152) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60632, Q61438 | Gene names | Nr2f1, Erbal3, Tfcoup1 | |||
|
Domain Architecture |
|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.024853 (rank : 109) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.015550 (rank : 131) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.018770 (rank : 124) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.030696 (rank : 88) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.031368 (rank : 86) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.006937 (rank : 151) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.026681 (rank : 107) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.054811 (rank : 43) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.052127 (rank : 47) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.021827 (rank : 116) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.051604 (rank : 48) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.028989 (rank : 102) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.032603 (rank : 84) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.032647 (rank : 82) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.029840 (rank : 96) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TULP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.011904 (rank : 137) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00295 | Gene names | TULP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 2 (Tubby-like protein 2). | |||||
|
CCD93_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.032113 (rank : 85) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.030628 (rank : 90) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.030456 (rank : 91) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.004370 (rank : 158) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.004741 (rank : 153) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.061859 (rank : 35) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
POF1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.029355 (rank : 100) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
RAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.011444 (rank : 138) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWZ1, O70452, O88391, Q3TGU1, Q3UG66, Q9QWZ3 | Gene names | Rad1, Rec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD1 (EC 3.1.11.2) (DNA repair exonuclease rad1 homolog) (Rad1-like DNA damage checkpoint protein) (mRAD1). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.023108 (rank : 113) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SCEL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.010924 (rank : 141) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.013940 (rank : 134) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
VIME_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.017269 (rank : 128) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P08670, Q15867, Q15868, Q15869, Q6LER9, Q8N850, Q96ML2, Q9NTM3 | Gene names | VIM | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
BMPR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.000213 (rank : 162) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35607 | Gene names | Bmpr2 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II) (BRK-3). | |||||
|
CCDC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.024159 (rank : 112) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D6Y1 | Gene names | Ccdc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 3 precursor. | |||||
|
CING_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.030633 (rank : 89) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
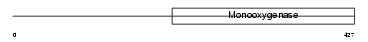
COQ6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.008628 (rank : 147) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1S0 | Gene names | Coq6 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquinone biosynthesis monooxygenase COQ6 (EC 1.14.13.-). | |||||
|
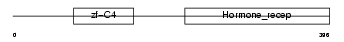
COT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.004222 (rank : 159) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10589 | Gene names | NR2F1, EAR3, ERBAL3, TFCOUP1 | |||
|
Domain Architecture |
|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I) (V-ERBA-related protein EAR-3). | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.011266 (rank : 139) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.027313 (rank : 105) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
INT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.007493 (rank : 148) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P4S8, Q80UQ7, Q91Z01, Q9CTF7 | Gene names | Ints1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.011918 (rank : 136) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.028328 (rank : 104) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KS6C1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.001927 (rank : 160) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
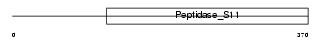
LACTB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.009509 (rank : 144) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |
|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
MPP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.021357 (rank : 117) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.018557 (rank : 125) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.018335 (rank : 126) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.015093 (rank : 132) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
NUP88_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.019035 (rank : 123) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99567, Q9BWE5 | Gene names | NUP88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup88 (Nucleoporin Nup88) (88 kDa nuclear pore complex protein). | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.068452 (rank : 34) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.031343 (rank : 87) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.030186 (rank : 94) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
POF1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.032639 (rank : 83) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K4L4, Q8C3A2 | Gene names | Pof1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.006948 (rank : 150) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RARG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.004720 (rank : 154) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
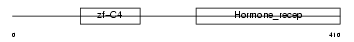
|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.004713 (rank : 155) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
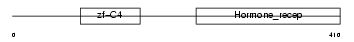
|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.004694 (rank : 156) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.004684 (rank : 157) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
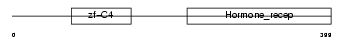
|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.011184 (rank : 140) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.024680 (rank : 110) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RMI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.008814 (rank : 145) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9A7, Q5SQG8, Q5SQG9, Q6P1Q4, Q6PI89, Q7Z6L6 | Gene names | RMI1, C9orf76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RMI1 homolog. | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.038465 (rank : 68) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.017713 (rank : 127) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
SEN54_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.008725 (rank : 146) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z6J9, Q86WV3, Q86XE4, Q8N9H2 | Gene names | TSEN54, SEN54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54) (HsSen54). | |||||
|
STP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.019304 (rank : 122) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05952, Q9NZB0 | Gene names | TNP2 | |||
|
Domain Architecture |
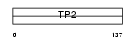
|
|||||
| Description | Nuclear transition protein 2 (TP-2) (TP2). | |||||
|
VIME_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.016708 (rank : 130) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P20152, O08704, Q8CCH1 | Gene names | Vim | |||
|
Domain Architecture |
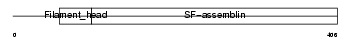
|
|||||
| Description | Vimentin. | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 162 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.994143 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L2_MOUSE
|
||||||
| NC score | 0.978058 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.977285 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.954631 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.940008 (rank : 6) | θ value | 9.45368e-175 (rank : 7) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.929786 (rank : 7) | θ value | 6.75929e-181 (rank : 6) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.883970 (rank : 8) | θ value | 9.45883e-58 (rank : 8) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| NC score | 0.458304 (rank : 9) | θ value | 7.09661e-13 (rank : 9) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| NC score | 0.445664 (rank : 10) | θ value | 6.00763e-12 (rank : 10) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.432395 (rank : 11) | θ value | 2.52405e-10 (rank : 11) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.410810 (rank : 12) | θ value | 1.06045e-08 (rank : 12) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.145236 (rank : 13) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.136462 (rank : 14) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.136433 (rank : 15) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA3_MOUSE
|
||||||
| NC score | 0.134827 (rank : 16) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.131771 (rank : 17) | θ value | 0.0148317 (rank : 16) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.121276 (rank : 18) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.120604 (rank : 19) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.120032 (rank : 20) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.113470 (rank : 21) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.101843 (rank : 22) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.101542 (rank : 23) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.101111 (rank : 24) | θ value | 0.00035302 (rank : 13) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.100948 (rank : 25) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.090692 (rank : 26) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.089154 (rank : 27) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.084870 (rank : 28) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.082732 (rank : 29) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.078498 (rank : 30) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.072528 (rank : 31) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.070884 (rank : 32) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.068903 (rank : 33) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.068452 (rank : 34) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.061859 (rank : 35) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
VAPB_HUMAN
|
||||||
| NC score | 0.061214 (rank : 36) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95292, O95293, Q9P0H0 | Gene names | VAPB | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B/C (VAMP- associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.060236 (rank : 37) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.058454 (rank : 38) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.057977 (rank : 39) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.056305 (rank : 40) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.055499 (rank : 41) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.055218 (rank : 42) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.054811 (rank : 43) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.053452 (rank : 44) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.052545 (rank : 45) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
VAPB_MOUSE
|
||||||
| NC score | 0.052145 (rank : 46) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QY76 | Gene names | Vapb | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B (VAMP- associated protein B) (VAMP-associated protein 33b) (VAMP-B) (VAP-B). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.052127 (rank : 47) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.051604 (rank : 48) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.050523 (rank : 49) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.047936 (rank : 50) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.047022 (rank : 51) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.046346 (rank : 52) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
THAP6_HUMAN
|
||||||
| NC score | 0.046031 (rank : 53) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.045559 (rank : 54) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.044151 (rank : 55) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.042918 (rank : 56) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.042466 (rank : 57) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.042410 (rank : 58) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.041946 (rank : 59) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
VAPA_HUMAN
|
||||||
| NC score | 0.041747 (rank : 60) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P0L0, O75453, Q5U0E7, Q9UBZ2 | Gene names | VAPA, VAP33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
VAPA_MOUSE
|
||||||
| NC score | 0.041486 (rank : 61) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV55, Q3TJM1, Q9QY77 | Gene names | Vapa, Vap33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.040987 (rank : 62) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.040445 (rank : 63) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.040248 (rank : 64) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.039427 (rank : 65) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.039357 (rank : 66) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.039018 (rank : 67) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.038465 (rank : 68) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.038430 (rank : 69) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.037667 (rank : 70) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.037353 (rank : 71) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.036827 (rank : 72) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.036615 (rank : 73) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.036294 (rank : 74) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MACOI_MOUSE
|
||||||
| NC score | 0.036252 (rank : 75) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.035223 (rank : 76) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.034019 (rank : 77) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.033882 (rank : 78) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.033706 (rank : 79) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.032994 (rank : 80) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.032675 (rank : 81) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.032647 (rank : 82) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
POF1B_MOUSE
|
||||||
| NC score | 0.032639 (rank : 83) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K4L4, Q8C3A2 | Gene names | Pof1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B. | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.032603 (rank : 84) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
CCD93_HUMAN
|
||||||
| NC score | 0.032113 (rank : 85) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.031368 (rank : 86) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.031343 (rank : 87) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.030696 (rank : 88) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.030633 (rank : 89) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.030628 (rank : 90) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.030456 (rank : 91) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.030362 (rank : 92) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.030276 (rank : 93) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.030186 (rank : 94) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SBDS_HUMAN
|
||||||
| NC score | 0.029925 (rank : 95) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3A5, Q96FX0, Q9NV53 | Gene names | SBDS | |||
|
Domain Architecture |

|
|||||
| Description | Shwachman-Bodian-Diamond syndrome protein. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.029840 (rank : 96) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.029686 (rank : 97) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.029584 (rank : 98) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.029562 (rank : 99) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
POF1B_HUMAN
|
||||||
| NC score | 0.029355 (rank : 100) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.029302 (rank : 101) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.028989 (rank : 102) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.028531 (rank : 103) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.028328 (rank : 104) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.027313 (rank : 105) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
SBDS_MOUSE
|
||||||
| NC score | 0.027233 (rank : 106) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70122, Q3TG78, Q9CR18 | Gene names | Sbds | |||
|
Domain Architecture |

|
|||||
| Description | Shwachman-Bodian-Diamond syndrome protein homolog (Protein 22A3). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.026681 (rank : 107) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.026344 (rank : 108) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.024853 (rank : 109) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.024680 (rank : 110) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.024271 (rank : 111) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCDC3_MOUSE
|
||||||
| NC score | 0.024159 (rank : 112) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D6Y1 | Gene names | Ccdc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 3 precursor. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.023108 (rank : 113) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
NSDHL_MOUSE
|
||||||
| NC score | 0.023050 (rank : 114) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1J0, O55109 | Gene names | Nsdhl | |||
|
Domain Architecture |

|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.021830 (rank : 115) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.021827 (rank : 116) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MPP4_HUMAN
|
||||||
| NC score | 0.021357 (rank : 117) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.021060 (rank : 118) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.020221 (rank : 119) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.020094 (rank : 120) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.019915 (rank : 121) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
STP2_HUMAN
|
||||||
| NC score | 0.019304 (rank : 122) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05952, Q9NZB0 | Gene names | TNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transition protein 2 (TP-2) (TP2). | |||||
|
NUP88_HUMAN
|
||||||
| NC score | 0.019035 (rank : 123) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99567, Q9BWE5 | Gene names | NUP88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup88 (Nucleoporin Nup88) (88 kDa nuclear pore complex protein). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.018770 (rank : 124) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.018557 (rank : 125) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.018335 (rank : 126) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.017713 (rank : 127) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
VIME_HUMAN
|
||||||
| NC score | 0.017269 (rank : 128) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P08670, Q15867, Q15868, Q15869, Q6LER9, Q8N850, Q96ML2, Q9NTM3 | Gene names | VIM | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.016925 (rank : 129) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
VIME_MOUSE
|
||||||
| NC score | 0.016708 (rank : 130) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P20152, O08704, Q8CCH1 | Gene names | Vim | |||
|
Domain Architecture |

|
|||||
| Description | Vimentin. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.015550 (rank : 131) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.015093 (rank : 132) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.014407 (rank : 133) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.013940 (rank : 134) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
OTOF_HUMAN
|
||||||
| NC score | 0.012035 (rank : 135) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.011918 (rank : 136) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
TULP2_HUMAN
|
||||||
| NC score | 0.011904 (rank : 137) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00295 | Gene names | TULP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 2 (Tubby-like protein 2). | |||||
|
RAD1_MOUSE
|
||||||
| NC score | 0.011444 (rank : 138) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWZ1, O70452, O88391, Q3TGU1, Q3UG66, Q9QWZ3 | Gene names | Rad1, Rec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD1 (EC 3.1.11.2) (DNA repair exonuclease rad1 homolog) (Rad1-like DNA damage checkpoint protein) (mRAD1). | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.011266 (rank : 139) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.011184 (rank : 140) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
SCEL_MOUSE
|
||||||
| NC score | 0.010924 (rank : 141) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.010868 (rank : 142) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.009828 (rank : 143) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LACTB_HUMAN
|
||||||
| NC score | 0.009509 (rank : 144) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
RMI1_HUMAN
|
||||||
| NC score | 0.008814 (rank : 145) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9A7, Q5SQG8, Q5SQG9, Q6P1Q4, Q6PI89, Q7Z6L6 | Gene names | RMI1, C9orf76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RMI1 homolog. | |||||
|
SEN54_HUMAN
|
||||||
| NC score | 0.008725 (rank : 146) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z6J9, Q86WV3, Q86XE4, Q8N9H2 | Gene names | TSEN54, SEN54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54) (HsSen54). | |||||
|
COQ6_MOUSE
|
||||||
| NC score | 0.008628 (rank : 147) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1S0 | Gene names | Coq6 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinone biosynthesis monooxygenase COQ6 (EC 1.14.13.-). | |||||
|
INT1_MOUSE
|
||||||
| NC score | 0.007493 (rank : 148) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P4S8, Q80UQ7, Q91Z01, Q9CTF7 | Gene names | Ints1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.007305 (rank : 149) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.006948 (rank : 150) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.006937 (rank : 151) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
COT1_MOUSE
|
||||||
| NC score | 0.004868 (rank : 152) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60632, Q61438 | Gene names | Nr2f1, Erbal3, Tfcoup1 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.004741 (rank : 153) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
RARG1_HUMAN
|
||||||
| NC score | 0.004720 (rank : 154) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG1_MOUSE
|
||||||
| NC score | 0.004713 (rank : 155) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG2_HUMAN
|
||||||
| NC score | 0.004694 (rank : 156) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARG2_MOUSE
|
||||||
| NC score | 0.004684 (rank : 157) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.004370 (rank : 158) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
COT1_HUMAN
|
||||||
| NC score | 0.004222 (rank : 159) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10589 | Gene names | NR2F1, EAR3, ERBAL3, TFCOUP1 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I) (V-ERBA-related protein EAR-3). | |||||
|
KS6C1_HUMAN
|
||||||
| NC score | 0.001927 (rank : 160) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96S38, Q8TDD3, Q9NSF4, Q9UL66 | Gene names | RPS6KC1, RPK118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase) (Ribosomal S6 kinase-like protein with two PSK domains 118 kDa protein) (SPHK1-binding protein). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.001191 (rank : 161) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
BMPR2_MOUSE
|
||||||
| NC score | 0.000213 (rank : 162) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 162 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35607 | Gene names | Bmpr2 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II) (BRK-3). | |||||