Please be patient as the page loads
|
Z3H7A_HUMAN
|
||||||
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
Z3H7A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.941103 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 3) | NC score | 0.469013 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 4) | NC score | 0.469034 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
OM34_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 5) | NC score | 0.493489 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 6) | NC score | 0.445519 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 7) | NC score | 0.459205 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
PR285_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 8) | NC score | 0.170864 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 9) | NC score | 0.443271 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 10) | NC score | 0.464821 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
OM34_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 11) | NC score | 0.490975 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 12) | NC score | 0.418119 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
PPID_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 13) | NC score | 0.263078 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 14) | NC score | 0.419231 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 15) | NC score | 0.436450 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
PPID_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 16) | NC score | 0.246675 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 17) | NC score | 0.447330 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 18) | NC score | 0.382090 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 19) | NC score | 0.400811 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 20) | NC score | 0.282811 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
TTC12_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.383904 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 22) | NC score | 0.392747 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 23) | NC score | 0.280248 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 24) | NC score | 0.293131 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.376870 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 26) | NC score | 0.389092 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.288717 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 28) | NC score | 0.212141 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.211485 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 30) | NC score | 0.294889 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
TTC4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.377401 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
ZN533_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 32) | NC score | 0.133673 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
ZN533_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 33) | NC score | 0.134145 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 34) | NC score | 0.124180 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 35) | NC score | 0.361742 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.295888 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.294444 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
ZN659_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 38) | NC score | 0.133418 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6B1 | Gene names | ZNF659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 659. | |||||
|
AIP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.214361 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.086236 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.093781 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 0.125558 (rank : 42) | NC score | 0.112189 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 0.125558 (rank : 43) | NC score | 0.111240 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
PPP5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 44) | NC score | 0.232032 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 45) | NC score | 0.117557 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.123929 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
PPP5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.233039 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.377286 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.089359 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.392369 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.054668 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.278888 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.032208 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PIAS2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.034594 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.039479 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
PIAS2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.033719 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5D8, O54987, Q8C384, Q8CDQ8, Q8K208, Q99JX5, Q9D5W7, Q9QZ63 | Gene names | Pias2, Miz1, Piasx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.006777 (rank : 129) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SP110_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.031079 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TTC4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.352689 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
AIP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.194429 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.166095 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
MOD5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.047148 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |
|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
PRDM4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.002436 (rank : 131) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.018109 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.095163 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
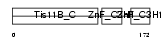
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.093895 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
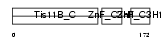
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
ZN518_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.011662 (rank : 126) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
ZN365_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.025627 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.015441 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.016118 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.013731 (rank : 122) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.075488 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
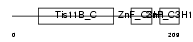
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.077900 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
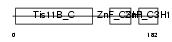
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.024459 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.039820 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.009182 (rank : 127) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RN169_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.023354 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.017467 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.141461 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.353318 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
UBL4A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.025429 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11441 | Gene names | UBL4A, DXS254E, GDX, UBL4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 4A (Ubiquitin-like protein GDX). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.000475 (rank : 134) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.035615 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.019952 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.015396 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.024360 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.039439 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
SPHK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.018923 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.034047 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
APR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.018568 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PGD0, Q3UAH4, Q810Q3, Q9DD14 | Gene names | Apr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-related protein 3 precursor (APR-3). | |||||
|
CLUL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.019300 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15846 | Gene names | CLUL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-like protein 1 precursor (Retinal-specific clusterin-like protein). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.020601 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.020545 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.166447 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.018083 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.019707 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.013635 (rank : 123) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.036345 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.014561 (rank : 121) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
PRDM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.000765 (rank : 133) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.012971 (rank : 124) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.007546 (rank : 128) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.005831 (rank : 130) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.002114 (rank : 132) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
VPS54_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.012412 (rank : 125) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Q0, Q5VIR5, Q86YF7, Q8N6G3, Q9NPV0, Q9NT07, Q9NUJ0 | Gene names | VPS54, HCC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p) (Hepatocellular carcinoma protein 8). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.197752 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.187387 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
APC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.066165 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.064278 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
BBS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.108181 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.105819 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.088828 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.150896 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.270302 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.288324 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
F10A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.161530 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
F10A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.175247 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
F10A4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.178094 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.161174 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.105105 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.102708 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.089424 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.078124 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
K0372_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.121594 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.120944 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.142462 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SUGT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.122498 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |
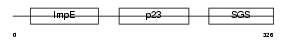
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
SUGT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.154265 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |
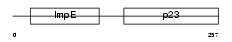
|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.081674 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
TTC13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.134740 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.110337 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.097389 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.110151 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.179148 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
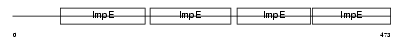
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.941103 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.493489 (rank : 3) | θ value | 1.53129e-07 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.490975 (rank : 4) | θ value | 9.92553e-07 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.469034 (rank : 5) | θ value | 4.0297e-08 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.469013 (rank : 6) | θ value | 2.13673e-09 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.464821 (rank : 7) | θ value | 7.59969e-07 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.459205 (rank : 8) | θ value | 4.45536e-07 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.447330 (rank : 9) | θ value | 1.43324e-05 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.445519 (rank : 10) | θ value | 3.41135e-07 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.443271 (rank : 11) | θ value | 7.59969e-07 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.436450 (rank : 12) | θ value | 4.92598e-06 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.419231 (rank : 13) | θ value | 3.77169e-06 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.418119 (rank : 14) | θ value | 1.69304e-06 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.400811 (rank : 15) | θ value | 0.000602161 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.392747 (rank : 16) | θ value | 0.00509761 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.392369 (rank : 17) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.389092 (rank : 18) | θ value | 0.0148317 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.383904 (rank : 19) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.382090 (rank : 20) | θ value | 0.00035302 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.377401 (rank : 21) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.377286 (rank : 22) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.376870 (rank : 23) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.361742 (rank : 24) | θ value | 0.0736092 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.353318 (rank : 25) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.352689 (rank : 26) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.295888 (rank : 27) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.294889 (rank : 28) | θ value | 0.0330416 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.294444 (rank : 29) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.293131 (rank : 30) | θ value | 0.0113563 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.288717 (rank : 31) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.288324 (rank : 32) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.282811 (rank : 33) | θ value | 0.00134147 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.280248 (rank : 34) | θ value | 0.00869519 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.278888 (rank : 35) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.270302 (rank : 36) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.263078 (rank : 37) | θ value | 2.21117e-06 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.246675 (rank : 38) | θ value | 1.09739e-05 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.233039 (rank : 39) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
PPP5_MOUSE
|
||||||
| NC score | 0.232032 (rank : 40) | θ value | 0.125558 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60676, O35299 | Gene names | Ppp5c | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PPT). | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.214361 (rank : 41) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.212141 (rank : 42) | θ value | 0.0193708 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.211485 (rank : 43) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.197752 (rank : 44) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.194429 (rank : 45) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.187387 (rank : 46) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.179148 (rank : 47) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.178094 (rank : 48) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
F10A1_MOUSE
|
||||||
| NC score | 0.175247 (rank : 49) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99L47, Q4FJT4 | Gene names | St13, Fam10a1, Hip | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Protein ST13 homolog) (Protein FAM10A1). | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.170864 (rank : 50) | θ value | 5.81887e-07 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.166447 (rank : 51) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.166095 (rank : 52) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.161530 (rank : 53) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.161174 (rank : 54) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
SUGT1_MOUSE
|
||||||
| NC score | 0.154265 (rank : 55) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CX34, Q3UF88, Q9CRE7, Q9D8M6 | Gene names | Sugt1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog. | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.150896 (rank : 56) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.142462 (rank : 57) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.141461 (rank : 58) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.134740 (rank : 59) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
ZN533_MOUSE
|
||||||
| NC score | 0.134145 (rank : 60) | θ value | 0.0563607 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
ZN533_HUMAN
|
||||||
| NC score | 0.133673 (rank : 61) | θ value | 0.0563607 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
ZN659_HUMAN
|
||||||
| NC score | 0.133418 (rank : 62) | θ value | 0.0961366 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6B1 | Gene names | ZNF659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 659. | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.124180 (rank : 63) | θ value | 0.0736092 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.123929 (rank : 64) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
SUGT1_HUMAN
|
||||||
| NC score | 0.122498 (rank : 65) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y2Z0, Q5JAK5, Q6VXY6 | Gene names | SUGT1 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of G2 allele of SKP1 homolog (Sgt1) (Putative 40-6-3 protein). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.121594 (rank : 66) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.120944 (rank : 67) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.117557 (rank : 68) | θ value | 0.125558 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.112189 (rank : 69) | θ value | 0.125558 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.111240 (rank : 70) | θ value | 0.125558 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.110337 (rank : 71) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.110151 (rank : 72) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.108181 (rank : 73) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.105819 (rank : 74) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.105105 (rank : 75) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.102708 (rank : 76) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.097389 (rank : 77) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.095163 (rank : 78) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.093895 (rank : 79) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.093781 (rank : 80) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.089424 (rank : 81) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.089359 (rank : 82) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.088828 (rank : 83) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.086236 (rank : 84) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.081674 (rank : 85) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.078124 (rank : 86) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.077900 (rank : 87) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.075488 (rank : 88) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
APC7_HUMAN
|
||||||
| NC score | 0.066165 (rank : 89) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
APC7_MOUSE
|
||||||
| NC score | 0.064278 (rank : 90) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.054668 (rank : 91) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
MOD5_HUMAN
|
||||||
| NC score | 0.047148 (rank : 92) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3H1, Q6IAC9, Q96FJ3, Q96L45, Q9NXT7 | Gene names | TRIT1, IPT, MOD5 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT) (hGRO1). | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.039820 (rank : 93) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.039479 (rank : 94) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.039439 (rank : 95) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.036345 (rank : 96) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.035615 (rank : 97) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PIAS2_HUMAN
|
||||||
| NC score | 0.034594 (rank : 98) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.034047 (rank : 99) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PIAS2_MOUSE
|
||||||
| NC score | 0.033719 (rank : 100) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5D8, O54987, Q8C384, Q8CDQ8, Q8K208, Q99JX5, Q9D5W7, Q9QZ63 | Gene names | Pias2, Miz1, Piasx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.032208 (rank : 101) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.031079 (rank : 102) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
ZN365_HUMAN
|
||||||
| NC score | 0.025627 (rank : 103) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
UBL4A_HUMAN
|
||||||
| NC score | 0.025429 (rank : 104) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11441 | Gene names | UBL4A, DXS254E, GDX, UBL4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 4A (Ubiquitin-like protein GDX). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.024459 (rank : 105) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.024360 (rank : 106) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.023354 (rank : 107) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.020601 (rank : 108) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.020545 (rank : 109) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.019952 (rank : 110) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.019707 (rank : 111) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
CLUL1_HUMAN
|
||||||
| NC score | 0.019300 (rank : 112) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15846 | Gene names | CLUL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-like protein 1 precursor (Retinal-specific clusterin-like protein). | |||||
|
SPHK2_MOUSE
|
||||||
| NC score | 0.018923 (rank : 113) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
APR3_MOUSE
|
||||||
| NC score | 0.018568 (rank : 114) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PGD0, Q3UAH4, Q810Q3, Q9DD14 | Gene names | Apr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-related protein 3 precursor (APR-3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.018109 (rank : 115) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.018083 (rank : 116) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.017467 (rank : 117) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.016118 (rank : 118) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.015441 (rank : 119) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.015396 (rank : 120) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.014561 (rank : 121) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.013731 (rank : 122) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.013635 (rank : 123) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.012971 (rank : 124) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
VPS54_HUMAN
|
||||||
| NC score | 0.012412 (rank : 125) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Q0, Q5VIR5, Q86YF7, Q8N6G3, Q9NPV0, Q9NT07, Q9NUJ0 | Gene names | VPS54, HCC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p) (Hepatocellular carcinoma protein 8). | |||||
|
ZN518_HUMAN
|
||||||
| NC score | 0.011662 (rank : 126) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.009182 (rank : 127) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.007546 (rank : 128) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.006777 (rank : 129) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.005831 (rank : 130) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
PRDM4_HUMAN
|
||||||
| NC score | 0.002436 (rank : 131) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.002114 (rank : 132) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
PRDM4_MOUSE
|
||||||
| NC score | 0.000765 (rank : 133) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.000475 (rank : 134) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||