Please be patient as the page loads
|
SF04_MOUSE
|
||||||
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SF04_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.980317 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 3) | NC score | 0.527697 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 4) | NC score | 0.533027 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 5) | NC score | 0.352694 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 6) | NC score | 0.350572 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.332425 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 8) | NC score | 0.322113 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 9) | NC score | 0.322244 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 10) | NC score | 0.343827 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 11) | NC score | 0.342777 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 12) | NC score | 0.326247 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 13) | NC score | 0.321679 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
GPTC2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.302690 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.296051 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 16) | NC score | 0.247849 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 17) | NC score | 0.242833 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
K0082_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.180042 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.172262 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.032044 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.053053 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.102530 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.138984 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.029833 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.043580 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
BAT4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.165118 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
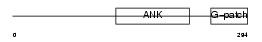
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
HHEX_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.022931 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43120 | Gene names | Hhex, Prhx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.033857 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
ROBO1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.019657 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.168211 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
BAT4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.166529 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
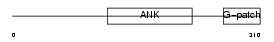
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.026857 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.086284 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.064062 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.040778 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.224224 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
PINX1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.222762 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.017822 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
AN32E_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.024540 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
EBP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.042562 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D903, Q3TKB5 | Gene names | Ebna1bp2, Ebp2 | |||
|
Domain Architecture |
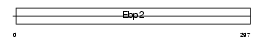
|
|||||
| Description | Probable rRNA-processing protein EBP2. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.046351 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.022691 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.024984 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.150866 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.124687 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
K0427_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.034794 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PEE2 | Gene names | Gm672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0427 homolog. | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.023458 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.041168 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.046655 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.043945 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
HHEX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.017403 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03014 | Gene names | HHEX, HEX, PRH, PRHX | |||
|
Domain Architecture |
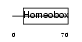
|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.040283 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.040473 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
SPT6H_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.045233 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.135939 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.033828 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.018763 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.025713 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
SPT6H_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.044228 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
BFAR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.023689 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.009773 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.031447 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.030373 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.022867 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
FOXJ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.007299 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
SFR16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.039001 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.005181 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.005913 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.038314 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.018149 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.032738 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
DLGP3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.022442 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.022083 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.027499 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.031067 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.039335 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.020190 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.020269 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.014931 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.038330 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ST2A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.013732 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50236 | Gene names | Sult2a2, Sta2, Sth2 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt sulfotransferase 2 (EC 2.8.2.14) (Hydroxysteroid sulfotransferase) (ST). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.023318 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.016230 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.084999 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.011953 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
LEUK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.031959 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.021955 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.066073 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.021506 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.030187 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.025071 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.015583 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.003490 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.004241 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SYIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.015321 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.111454 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.054161 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.980317 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.533027 (rank : 3) | θ value | 2.88119e-14 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.527697 (rank : 4) | θ value | 3.07829e-16 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.352694 (rank : 5) | θ value | 5.62301e-10 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.350572 (rank : 6) | θ value | 6.21693e-09 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.343827 (rank : 7) | θ value | 3.19293e-05 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.342777 (rank : 8) | θ value | 3.19293e-05 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.332425 (rank : 9) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.326247 (rank : 10) | θ value | 5.44631e-05 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.322244 (rank : 11) | θ value | 1.87187e-05 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.322113 (rank : 12) | θ value | 1.87187e-05 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.321679 (rank : 13) | θ value | 0.000158464 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.302690 (rank : 14) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.296051 (rank : 15) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.247849 (rank : 16) | θ value | 0.00390308 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.242833 (rank : 17) | θ value | 0.00665767 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.224224 (rank : 18) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.222762 (rank : 19) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.180042 (rank : 20) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.172262 (rank : 21) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.168211 (rank : 22) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.166529 (rank : 23) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.165118 (rank : 24) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.150866 (rank : 25) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.138984 (rank : 26) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.135939 (rank : 27) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.124687 (rank : 28) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.111454 (rank : 29) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.102530 (rank : 30) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.086284 (rank : 31) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.084999 (rank : 32) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.066073 (rank : 33) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.064062 (rank : 34) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.054161 (rank : 35) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.053053 (rank : 36) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.046655 (rank : 37) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.046351 (rank : 38) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SPT6H_MOUSE
|
||||||
| NC score | 0.045233 (rank : 39) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
SPT6H_HUMAN
|
||||||
| NC score | 0.044228 (rank : 40) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.043945 (rank : 41) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.043580 (rank : 42) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
EBP2_MOUSE
|
||||||
| NC score | 0.042562 (rank : 43) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D903, Q3TKB5 | Gene names | Ebna1bp2, Ebp2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable rRNA-processing protein EBP2. | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.041168 (rank : 44) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.040778 (rank : 45) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.040473 (rank : 46) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.040283 (rank : 47) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.039335 (rank : 48) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.039001 (rank : 49) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.038330 (rank : 50) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.038314 (rank : 51) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
K0427_MOUSE
|
||||||
| NC score | 0.034794 (rank : 52) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PEE2 | Gene names | Gm672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0427 homolog. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.033857 (rank : 53) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.033828 (rank : 54) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.032738 (rank : 55) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.032044 (rank : 56) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.031959 (rank : 57) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.031447 (rank : 58) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.031067 (rank : 59) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.030373 (rank : 60) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.030187 (rank : 61) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.029833 (rank : 62) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.027499 (rank : 63) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.026857 (rank : 64) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.025713 (rank : 65) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.025071 (rank : 66) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.024984 (rank : 67) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
AN32E_HUMAN
|
||||||
| NC score | 0.024540 (rank : 68) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
BFAR_MOUSE
|
||||||
| NC score | 0.023689 (rank : 69) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.023458 (rank : 70) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.023318 (rank : 71) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
HHEX_MOUSE
|
||||||
| NC score | 0.022931 (rank : 72) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43120 | Gene names | Hhex, Prhx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.022867 (rank : 73) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.022691 (rank : 74) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
DLGP3_MOUSE
|
||||||
| NC score | 0.022442 (rank : 75) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.022083 (rank : 76) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.021955 (rank : 77) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.021506 (rank : 78) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.020269 (rank : 79) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.020190 (rank : 80) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
ROBO1_MOUSE
|
||||||
| NC score | 0.019657 (rank : 81) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.018763 (rank : 82) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.018149 (rank : 83) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | 0.017822 (rank : 84) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
HHEX_HUMAN
|
||||||
| NC score | 0.017403 (rank : 85) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03014 | Gene names | HHEX, HEX, PRH, PRHX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.016230 (rank : 86) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.015583 (rank : 87) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
SYIC_MOUSE
|
||||||
| NC score | 0.015321 (rank : 88) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.014931 (rank : 89) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
ST2A2_MOUSE
|
||||||
| NC score | 0.013732 (rank : 90) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50236 | Gene names | Sult2a2, Sta2, Sth2 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt sulfotransferase 2 (EC 2.8.2.14) (Hydroxysteroid sulfotransferase) (ST). | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.011953 (rank : 91) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.009773 (rank : 92) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FOXJ1_MOUSE
|
||||||
| NC score | 0.007299 (rank : 93) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.005913 (rank : 94) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.005181 (rank : 95) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.004241 (rank : 96) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.003490 (rank : 97) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||