Please be patient as the page loads
|
PINX1_MOUSE
|
||||||
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PINX1_MOUSE
|
||||||
| θ value | 2.59246e-148 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 1.10445e-98 (rank : 2) | NC score | 0.956712 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 3) | NC score | 0.417132 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 4) | NC score | 0.416269 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 5) | NC score | 0.123353 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 6) | NC score | 0.356837 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 7) | NC score | 0.356439 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.302813 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
GPTC2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 9) | NC score | 0.331677 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 10) | NC score | 0.323690 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 11) | NC score | 0.335690 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 12) | NC score | 0.294256 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 13) | NC score | 0.318259 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 14) | NC score | 0.316287 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.122457 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.115234 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.163780 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
VPK7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.266016 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.033593 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.276917 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.274897 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
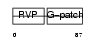
VPK10_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.264357 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.264324 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.265252 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.264650 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.266073 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.262885 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.260719 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.213778 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.055843 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
VPK3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.255842 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.042797 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.064167 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SPF45_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.175798 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
VPK11_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.252461 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.171963 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
K0082_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.216894 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
SF04_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.229804 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.222762 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
CS021_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.052765 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.039800 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.168648 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.133760 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
VPK4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.232750 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.232825 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
4EBP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.036715 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13541, Q6IBN3 | Gene names | EIF4EBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (eIF4E-binding protein 1) (Phosphorylated heat- and acid-stable protein regulated by insulin 1) (PHAS-I). | |||||
|
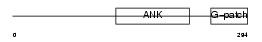
BAT4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.154238 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
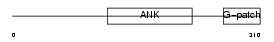
BAT4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.162358 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.113624 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.048281 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.170419 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZN512_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.027514 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96ME7, Q96JM0 | Gene names | ZNF512, KIAA1805 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 512. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.012087 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.048136 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.036696 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.039391 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.005871 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.016263 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
GPSM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.010530 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.019703 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.054823 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.018623 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
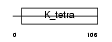
ZN238_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.000346 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99592, Q13397, Q5VU40, Q8N463 | Gene names | ZNF238, RP58, TAZ1, ZBTB18 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 238 (Transcriptional repressor RP58) (58 kDa repressor protein) (Zinc finger protein C2H2-171) (Translin-associated zinc finger protein 1) (TAZ-1) (Zinc finger and BTB domain-containing protein 18). | |||||
|
ZN238_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | -0.000019 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUK6 | Gene names | Znf238, Rp58, Zfp238 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 238 (Zfp-238) (Transcriptional repressor RP58) (58 kDa repressor protein). | |||||
|
CN118_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.052866 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
CN118_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.057454 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051383 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051182 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.056914 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RBM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.115182 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.59246e-148 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.956712 (rank : 2) | θ value | 1.10445e-98 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.417132 (rank : 3) | θ value | 9.92553e-07 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.416269 (rank : 4) | θ value | 1.87187e-05 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.356837 (rank : 5) | θ value | 9.29e-05 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.356439 (rank : 6) | θ value | 9.29e-05 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.335690 (rank : 7) | θ value | 0.00509761 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.331677 (rank : 8) | θ value | 0.00390308 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.323690 (rank : 9) | θ value | 0.00509761 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.318259 (rank : 10) | θ value | 0.00665767 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.316287 (rank : 11) | θ value | 0.00665767 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.302813 (rank : 12) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.294256 (rank : 13) | θ value | 0.00509761 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.276917 (rank : 14) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.274897 (rank : 15) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
VPK9_HUMAN
|
||||||
| NC score | 0.266073 (rank : 16) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK7_HUMAN
|
||||||
| NC score | 0.266016 (rank : 17) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| NC score | 0.265252 (rank : 18) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| NC score | 0.264650 (rank : 19) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK10_HUMAN
|
||||||
| NC score | 0.264357 (rank : 20) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| NC score | 0.264324 (rank : 21) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| NC score | 0.262885 (rank : 22) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| NC score | 0.260719 (rank : 23) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| NC score | 0.255842 (rank : 24) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| NC score | 0.252461 (rank : 25) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| NC score | 0.232825 (rank : 26) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK4_HUMAN
|
||||||
| NC score | 0.232750 (rank : 27) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.229804 (rank : 28) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.222762 (rank : 29) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.216894 (rank : 30) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.213778 (rank : 31) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.175798 (rank : 32) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.171963 (rank : 33) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.170419 (rank : 34) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.168648 (rank : 35) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.163780 (rank : 36) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.162358 (rank : 37) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.154238 (rank : 38) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.133760 (rank : 39) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.123353 (rank : 40) | θ value | 3.19293e-05 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.122457 (rank : 41) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.115234 (rank : 42) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.115182 (rank : 43) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.113624 (rank : 44) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.064167 (rank : 45) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CN118_MOUSE
|
||||||
| NC score | 0.057454 (rank : 46) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.056914 (rank : 47) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055843 (rank : 48) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.054823 (rank : 49) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CN118_HUMAN
|
||||||
| NC score | 0.052866 (rank : 50) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
CS021_MOUSE
|
||||||
| NC score | 0.052765 (rank : 51) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.051383 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.051182 (rank : 53) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.048281 (rank : 54) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.048136 (rank : 55) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.042797 (rank : 56) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.039800 (rank : 57) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.039391 (rank : 58) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
4EBP1_HUMAN
|
||||||
| NC score | 0.036715 (rank : 59) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13541, Q6IBN3 | Gene names | EIF4EBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (eIF4E-binding protein 1) (Phosphorylated heat- and acid-stable protein regulated by insulin 1) (PHAS-I). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.036696 (rank : 60) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.033593 (rank : 61) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
ZN512_HUMAN
|
||||||
| NC score | 0.027514 (rank : 62) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96ME7, Q96JM0 | Gene names | ZNF512, KIAA1805 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 512. | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.019703 (rank : 63) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.018623 (rank : 64) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.016263 (rank : 65) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.012087 (rank : 66) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
GPSM2_MOUSE
|
||||||
| NC score | 0.010530 (rank : 67) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.005871 (rank : 68) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ZN238_HUMAN
|
||||||
| NC score | 0.000346 (rank : 69) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99592, Q13397, Q5VU40, Q8N463 | Gene names | ZNF238, RP58, TAZ1, ZBTB18 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 238 (Transcriptional repressor RP58) (58 kDa repressor protein) (Zinc finger protein C2H2-171) (Translin-associated zinc finger protein 1) (TAZ-1) (Zinc finger and BTB domain-containing protein 18). | |||||
|
ZN238_MOUSE
|
||||||
| NC score | -0.000019 (rank : 70) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUK6 | Gene names | Znf238, Rp58, Zfp238 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 238 (Zfp-238) (Transcriptional repressor RP58) (58 kDa repressor protein). | |||||