Please be patient as the page loads
|
GPTC2_MOUSE
|
||||||
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GPTC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994415 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
CN118_MOUSE
|
||||||
| θ value | 3.26607e-42 (rank : 3) | NC score | 0.745964 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
CN118_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 4) | NC score | 0.720612 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 5) | NC score | 0.396695 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.395807 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 7) | NC score | 0.392827 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 8) | NC score | 0.389763 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 9) | NC score | 0.388514 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 10) | NC score | 0.338128 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 11) | NC score | 0.361032 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 12) | NC score | 0.345655 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 13) | NC score | 0.366059 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 14) | NC score | 0.320235 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 15) | NC score | 0.319159 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
SF04_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 16) | NC score | 0.302668 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.296051 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.242651 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 19) | NC score | 0.318018 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
PINX1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 20) | NC score | 0.323690 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.235738 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
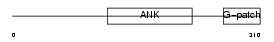
BAT4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.210514 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
VPK1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.239234 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 24) | NC score | 0.239605 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.235961 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
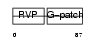
VPK10_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.232945 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.232876 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.233183 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
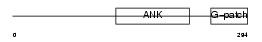
BAT4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 29) | NC score | 0.199921 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
VPK11_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.225374 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.223386 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.218367 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
MORC4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.043854 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
VPK4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.207972 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.208083 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.219014 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
K0082_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.139706 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MORC4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.041406 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.007148 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.021991 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
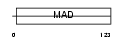
CT151_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.028422 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.011675 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.011961 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.101097 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.012707 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.009492 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
ADA25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.008054 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R159, Q9D4E4 | Gene names | Adam25 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 25 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 25) (Testase 2). | |||||
|
APOC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.033282 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02654 | Gene names | APOC1 | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein C-I precursor (Apo-CI) (ApoC-I). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.005831 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.052561 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.078349 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.001913 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.141128 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.008580 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.006385 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.009024 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.129811 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007272 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.009202 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.004954 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
ZBT7A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | -0.002406 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95365, O00456 | Gene names | ZBTB7A, FBI1, LRF, ZBTB7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 7A (Leukemia/lymphoma- related factor) (Factor that binds to inducer of short transcripts protein 1) (Factor binding IST protein 1) (FBI-1) (HIV-1 1st-binding protein 1) (TTF-I-interacting peptide 21) (TIP21). | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.057891 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
SPF45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.053286 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.054271 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.994415 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
CN118_MOUSE
|
||||||
| NC score | 0.745964 (rank : 3) | θ value | 3.26607e-42 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
CN118_HUMAN
|
||||||
| NC score | 0.720612 (rank : 4) | θ value | 3.06405e-32 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.396695 (rank : 5) | θ value | 1.38499e-08 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.395807 (rank : 6) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.392827 (rank : 7) | θ value | 1.17247e-07 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.389763 (rank : 8) | θ value | 7.59969e-07 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.388514 (rank : 9) | θ value | 7.59969e-07 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.366059 (rank : 10) | θ value | 1.87187e-05 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.361032 (rank : 11) | θ value | 1.43324e-05 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.345655 (rank : 12) | θ value | 1.43324e-05 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.338128 (rank : 13) | θ value | 8.40245e-06 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.323690 (rank : 14) | θ value | 0.00509761 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.320235 (rank : 15) | θ value | 0.000270298 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.319159 (rank : 16) | θ value | 0.000270298 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.318018 (rank : 17) | θ value | 0.00509761 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.302668 (rank : 18) | θ value | 0.00102713 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.296051 (rank : 19) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.242651 (rank : 20) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
VPK9_HUMAN
|
||||||
| NC score | 0.239605 (rank : 21) | θ value | 0.0148317 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| NC score | 0.239234 (rank : 22) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| NC score | 0.235961 (rank : 23) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.235738 (rank : 24) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
VPK5_HUMAN
|
||||||
| NC score | 0.233183 (rank : 25) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK10_HUMAN
|
||||||
| NC score | 0.232945 (rank : 26) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| NC score | 0.232876 (rank : 27) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| NC score | 0.225374 (rank : 28) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK7_HUMAN
|
||||||
| NC score | 0.223386 (rank : 29) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| NC score | 0.219014 (rank : 30) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| NC score | 0.218367 (rank : 31) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.210514 (rank : 32) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
VPK6_HUMAN
|
||||||
| NC score | 0.208083 (rank : 33) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK4_HUMAN
|
||||||
| NC score | 0.207972 (rank : 34) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.199921 (rank : 35) | θ value | 0.0563607 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.141128 (rank : 36) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.139706 (rank : 37) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.129811 (rank : 38) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.101097 (rank : 39) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.078349 (rank : 40) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.057891 (rank : 41) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.054271 (rank : 42) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SPF45_HUMAN
|
||||||
| NC score | 0.053286 (rank : 43) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I25, Q96GY6 | Gene names | RBM17, SPF45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.052561 (rank : 44) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MORC4_MOUSE
|
||||||
| NC score | 0.043854 (rank : 45) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MORC4_HUMAN
|
||||||
| NC score | 0.041406 (rank : 46) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
APOC1_HUMAN
|
||||||
| NC score | 0.033282 (rank : 47) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02654 | Gene names | APOC1 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein C-I precursor (Apo-CI) (ApoC-I). | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.028422 (rank : 48) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.021991 (rank : 49) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.012707 (rank : 50) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.011961 (rank : 51) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.011675 (rank : 52) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.009492 (rank : 53) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.009202 (rank : 54) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.009024 (rank : 55) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.008580 (rank : 56) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
ADA25_MOUSE
|
||||||
| NC score | 0.008054 (rank : 57) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R159, Q9D4E4 | Gene names | Adam25 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 25 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 25) (Testase 2). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.007272 (rank : 58) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.007148 (rank : 59) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.006385 (rank : 60) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.005831 (rank : 61) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.004954 (rank : 62) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.001913 (rank : 63) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
ZBT7A_HUMAN
|
||||||
| NC score | -0.002406 (rank : 64) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95365, O00456 | Gene names | ZBTB7A, FBI1, LRF, ZBTB7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 7A (Leukemia/lymphoma- related factor) (Factor that binds to inducer of short transcripts protein 1) (Factor binding IST protein 1) (FBI-1) (HIV-1 1st-binding protein 1) (TTF-I-interacting peptide 21) (TIP21). | |||||