Please be patient as the page loads
|
CB027_HUMAN
|
||||||
| SwissProt Accessions | Q580R0, O43575, Q52M10, Q86XG2 | Gene names | C2orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf27. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CB027_HUMAN
|
||||||
| θ value | 3.81361e-83 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q580R0, O43575, Q52M10, Q86XG2 | Gene names | C2orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf27. | |||||
|
RGDSR_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 2) | NC score | 0.209783 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |
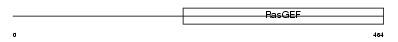
|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 3) | NC score | 0.081822 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 4) | NC score | 0.059658 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 5) | NC score | 0.059469 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 1.38821 (rank : 6) | NC score | 0.043060 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.094643 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.033582 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CISH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.034744 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSE2, Q9NS38, Q9Y5R1 | Gene names | CISH, G18 | |||
|
Domain Architecture |
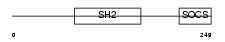
|
|||||
| Description | Cytokine-inducible SH2-containing protein (CIS) (CIS-1) (Suppressor of cytokine signaling) (SOCS) (Protein G18). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.061081 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.099646 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.083305 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.045259 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.038277 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PLIN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.038970 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGN5 | Gene names | Plin, Peri | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.046761 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.067713 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NPT2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.025363 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06495 | Gene names | SLC34A1, NPT2, SLC17A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent phosphate transport protein 2A (Sodium/phosphate cotransporter 2A) (Na(+)/Pi cotransporter 2A) (Sodium-phosphate transport protein 2A) (Na(+)-dependent phosphate cotransporter 2A) (NaPi-2a) (Solute carrier family 34 member 1) (NaPi-3). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.039465 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.042407 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
VCX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.149511 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H322, Q9P0H5 | Gene names | VCX2, VCX2R, VCXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 2 (VCX-B protein) (Variably charged protein X-B) (Variable charge protein on X with two repeats) (VCX-2r). | |||||
|
VCY1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.137775 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.008827 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.001993 (rank : 36) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.019445 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
CILP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.015788 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUL8, Q6NV88, Q8N4A6, Q8WV21 | Gene names | CILP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 2 precursor (CILP-2) [Contains: Cartilage intermediate layer protein 2 C1; Cartilage intermediate layer protein 2 C2]. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.011927 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
GIDRP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.025113 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.004969 (rank : 35) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.071285 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
GNDS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.069143 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
RGL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.056110 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NZL6, Q9HBY3, Q9HBY4, Q9NZL5, Q9UG43, Q9Y2G6 | Gene names | RGL1, KIAA0959, RGL | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.056262 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.055865 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15211, Q9Y3F3 | Gene names | RGL2, RAB2L | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.054276 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.052739 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CB027_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.81361e-83 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q580R0, O43575, Q52M10, Q86XG2 | Gene names | C2orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf27. | |||||
|
RGDSR_HUMAN
|
||||||
| NC score | 0.209783 (rank : 2) | θ value | 2.21117e-06 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZJ4 | Gene names | RGR | |||
|
Domain Architecture |

|
|||||
| Description | Ral-GDS-related protein (hRGR). | |||||
|
VCX2_HUMAN
|
||||||
| NC score | 0.149511 (rank : 3) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H322, Q9P0H5 | Gene names | VCX2, VCX2R, VCXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 2 (VCX-B protein) (Variably charged protein X-B) (Variable charge protein on X with two repeats) (VCX-2r). | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.137775 (rank : 4) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.099646 (rank : 5) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.094643 (rank : 6) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.083305 (rank : 7) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.081822 (rank : 8) | θ value | 0.00390308 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.071285 (rank : 9) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.069143 (rank : 10) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.067713 (rank : 11) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.061081 (rank : 12) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.059658 (rank : 13) | θ value | 0.0961366 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.059469 (rank : 14) | θ value | 1.06291 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
RGL1_MOUSE
|
||||||
| NC score | 0.056262 (rank : 15) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL1_HUMAN
|
||||||
| NC score | 0.056110 (rank : 16) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NZL6, Q9HBY3, Q9HBY4, Q9NZL5, Q9UG43, Q9Y2G6 | Gene names | RGL1, KIAA0959, RGL | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
RGL2_HUMAN
|
||||||
| NC score | 0.055865 (rank : 17) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15211, Q9Y3F3 | Gene names | RGL2, RAB2L | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGL2_MOUSE
|
||||||
| NC score | 0.054276 (rank : 18) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.052739 (rank : 19) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.046761 (rank : 20) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.045259 (rank : 21) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.043060 (rank : 22) | θ value | 1.38821 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.042407 (rank : 23) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.039465 (rank : 24) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PLIN_MOUSE
|
||||||
| NC score | 0.038970 (rank : 25) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGN5 | Gene names | Plin, Peri | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.038277 (rank : 26) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CISH_HUMAN
|
||||||
| NC score | 0.034744 (rank : 27) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSE2, Q9NS38, Q9Y5R1 | Gene names | CISH, G18 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine-inducible SH2-containing protein (CIS) (CIS-1) (Suppressor of cytokine signaling) (SOCS) (Protein G18). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.033582 (rank : 28) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
NPT2A_HUMAN
|
||||||
| NC score | 0.025363 (rank : 29) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06495 | Gene names | SLC34A1, NPT2, SLC17A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent phosphate transport protein 2A (Sodium/phosphate cotransporter 2A) (Na(+)/Pi cotransporter 2A) (Sodium-phosphate transport protein 2A) (Na(+)-dependent phosphate cotransporter 2A) (NaPi-2a) (Solute carrier family 34 member 1) (NaPi-3). | |||||
|
GIDRP_MOUSE
|
||||||
| NC score | 0.025113 (rank : 30) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.019445 (rank : 31) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
CILP2_HUMAN
|
||||||
| NC score | 0.015788 (rank : 32) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUL8, Q6NV88, Q8N4A6, Q8WV21 | Gene names | CILP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 2 precursor (CILP-2) [Contains: Cartilage intermediate layer protein 2 C1; Cartilage intermediate layer protein 2 C2]. | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.011927 (rank : 33) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.008827 (rank : 34) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.004969 (rank : 35) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
ZN710_HUMAN
|
||||||
| NC score | 0.001993 (rank : 36) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||