Please be patient as the page loads
|
SEM3D_MOUSE
|
||||||
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SEM3A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994718 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994398 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.990024 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3C_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.990626 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3C_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.990084 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999362 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
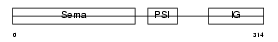
|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3D_MOUSE
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3E_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.992714 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM3E_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.992548 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3F_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.990227 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM3F_MOUSE
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.990777 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM3B_MOUSE
|
||||||
| θ value | 1.3161e-184 (rank : 12) | NC score | 0.990618 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM4C_MOUSE
|
||||||
| θ value | 2.30293e-96 (rank : 13) | NC score | 0.964212 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 2.01746e-92 (rank : 14) | NC score | 0.962044 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4D_HUMAN
|
||||||
| θ value | 3.0147e-88 (rank : 15) | NC score | 0.962159 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 5.14231e-88 (rank : 16) | NC score | 0.959720 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4C_HUMAN
|
||||||
| θ value | 7.42547e-87 (rank : 17) | NC score | 0.958793 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4G_HUMAN
|
||||||
| θ value | 1.65422e-86 (rank : 18) | NC score | 0.959607 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 2.16048e-86 (rank : 19) | NC score | 0.939010 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 2.91998e-83 (rank : 20) | NC score | 0.939672 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM4B_MOUSE
|
||||||
| θ value | 7.19216e-82 (rank : 21) | NC score | 0.953633 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 3.56943e-81 (rank : 22) | NC score | 0.955510 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4F_HUMAN
|
||||||
| θ value | 2.31364e-80 (rank : 23) | NC score | 0.959295 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |
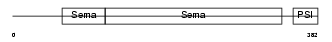
|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 2.82823e-78 (rank : 24) | NC score | 0.941245 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 1.18825e-76 (rank : 25) | NC score | 0.953505 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 5.8972e-76 (rank : 26) | NC score | 0.940214 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM4F_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 27) | NC score | 0.959449 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4A_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 28) | NC score | 0.952170 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 2.17053e-70 (rank : 29) | NC score | 0.940918 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 4.52582e-68 (rank : 30) | NC score | 0.926934 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM5B_MOUSE
|
||||||
| θ value | 1.6097e-65 (rank : 31) | NC score | 0.785423 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5B_HUMAN
|
||||||
| θ value | 6.11685e-65 (rank : 32) | NC score | 0.788445 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM5A_HUMAN
|
||||||
| θ value | 1.77973e-64 (rank : 33) | NC score | 0.783895 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM5A_MOUSE
|
||||||
| θ value | 1.50663e-63 (rank : 34) | NC score | 0.788583 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 35) | NC score | 0.934490 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 3.71096e-62 (rank : 36) | NC score | 0.932892 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM7A_HUMAN
|
||||||
| θ value | 4.70527e-49 (rank : 37) | NC score | 0.942386 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |
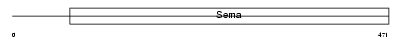
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM7A_MOUSE
|
||||||
| θ value | 1.85029e-45 (rank : 38) | NC score | 0.940053 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |
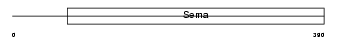
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
PLXA2_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 39) | NC score | 0.296638 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 40) | NC score | 0.295367 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA4_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 41) | NC score | 0.304137 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 42) | NC score | 0.327039 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 43) | NC score | 0.303465 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXC1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 44) | NC score | 0.308199 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.313570 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | -0.001014 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
PLXA3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 47) | NC score | 0.275198 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
IRPL2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 48) | NC score | 0.032537 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ERS6, Q9ER66 | Gene names | Il1rapl2 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 2 precursor (IL1RAPL-2-related protein) (TIGIRR-1). | |||||
|
CD79B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 49) | NC score | 0.055610 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15530 | Gene names | Cd79b, Igb | |||
|
Domain Architecture |
|
|||||
| Description | B-cell antigen receptor complex-associated protein beta-chain precursor (B-cell-specific glycoprotein B29) (Immunoglobulin- associated B29 protein) (IG-beta) (CD79b antigen). | |||||
|
IRPL2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.031601 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NP60, Q9NZN0 | Gene names | IL1RAPL2, IL1R9 | |||
|
Domain Architecture |
|
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 2 precursor (IL1RAPL-2-related protein) (Interleukin-1 receptor 9) (IL-1R9) (IL-1 receptor accessory protein-like 2) (Three immunoglobulin domain- containing IL-1 receptor-related 1) (TIGIRR-1). | |||||
|
TRI34_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.010253 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYJ4, Q9C016, Q9HCR0, Q9HCR1, Q9HCR2 | Gene names | TRIM34, IFP1, RNF21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 34 (Interferon-responsive finger protein 1) (RING finger protein 21). | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.281703 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.235954 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXA1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.272333 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.020986 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
CNTN4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.035930 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q69Z26, Q8BRT6, Q8CBD3 | Gene names | Cntn4, Kiaa3024 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
L1CAM_MOUSE
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.019377 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11627 | Gene names | L1cam, Caml1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
CNTN4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.037893 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
LRRN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.010100 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 508 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61809, Q3USY1, Q69ZI0 | Gene names | Lrrn1, Kiaa1497 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats neuronal protein 1 precursor (Neuronal leucine- rich repeat protein 1) (NLRR-1). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.028682 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
KV6E_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.012958 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01679 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region J539. | |||||
|
L1CAM_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.021034 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P32004, Q8TA87 | Gene names | L1CAM, CAML1, MIC5 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.262238 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
IRPL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.024172 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZN1, Q9UJ53 | Gene names | IL1RAPL1, OPHN4 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 1 precursor (IL1RAPL-1) (Oligophrenin-4) (Three immunoglobulin domain-containing IL-1 receptor-related 2) (TIGIRR-2). | |||||
|
IRPL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.024495 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59823 | Gene names | Il1rapl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 1 precursor (IL1RAPL-1). | |||||
|
KV6F_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.012407 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04940 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-VI region NQ2-17.4.1. | |||||
|
KV6G_MOUSE
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.012505 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04941 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-VI region NQ2-48.2.2. | |||||
|
KV6I_MOUSE
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.012575 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04943 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-VI region NQ6-8.3.1. | |||||
|
KV6J_MOUSE
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.012613 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04944 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-VI region NQ5-78.2.6. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.262715 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
AMGO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.015487 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86WK6, Q8IW71, Q9ULQ7 | Gene names | AMIGO1, ALI2, AMIGO, KIAA1163 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amphoterin-induced protein 1 precursor (AMIGO-1) (Alivin-2). | |||||
|
KV6B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.011899 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01676 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region XRPC 24. | |||||
|
KV6C_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.011994 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01677 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region TEPC 601/TEPC 191. | |||||
|
KV6D_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.011829 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01678 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region SAPC 10. | |||||
|
KV6H_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.011711 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04942 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-VI region NQ5-61.1.2. | |||||
|
LRRN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.009388 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UXK5, Q3LID5, Q8IYV5, Q9H8V1, Q9P231 | Gene names | LRRN1, KIAA1497 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats neuronal protein 1 precursor (Neuronal leucine- rich repeat protein 1) (NLRR-1). | |||||
|
PROX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.014718 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
PROX1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.014657 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | -0.001920 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
KV6A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.011010 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01675 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region XRPC 44. | |||||
|
CNTN5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.015319 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94779, O94780 | Gene names | CNTN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2) (hNB-2). | |||||
|
KV1W_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.007466 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04431 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-I region Walker precursor. | |||||
|
CNTN6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.016205 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.004861 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.000488 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.006803 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.008221 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CD79B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.038121 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40259, Q9BU06 | Gene names | CD79B, B29, IGB | |||
|
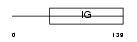
Domain Architecture |
|
|||||
| Description | B-cell antigen receptor complex-associated protein beta-chain precursor (B-cell-specific glycoprotein B29) (Immunoglobulin- associated B29 protein) (IG-beta) (CD79b antigen). | |||||
|
CEAM6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.007801 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P40199, Q14920 | Gene names | CEACAM6, NCA | |||
|
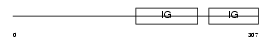
Domain Architecture |
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 6 precursor (Normal cross-reacting antigen) (Nonspecific crossreacting antigen) (CD66c antigen). | |||||
|
ITB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.001081 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
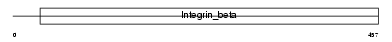
Domain Architecture |
|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.010301 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CILP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.041554 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q66K08, Q7TSS0, Q7TSS1, Q8BV01 | Gene names | Cilp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 1 precursor (CILP-1) [Contains: Cartilage intermediate layer protein 1 C1; Cartilage intermediate layer protein 1 C2]. | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.000464 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
CILP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.035681 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75339, Q6UW99, Q8IYI5 | Gene names | CILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 1 precursor (CILP-1) (Cartilage intermediate-layer protein) [Contains: Cartilage intermediate layer protein 1 C1; Cartilage intermediate layer protein 1 C2]. | |||||
|
KV2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.006204 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P01615 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region FR. | |||||
|
MERTK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | -0.001820 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12866, Q9HBB4 | Gene names | MERTK, MER | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
TMC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.002549 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI8 | Gene names | TMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1. | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.233415 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.236935 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PROP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.087012 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
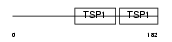
Domain Architecture |
|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.084607 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SPON1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.063124 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
SPON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.064588 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
UNC5A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.054932 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
UNC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058621 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.058594 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
UNC5C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053354 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
UNC5C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054302 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08747, Q8CD16 | Gene names | Unc5c, Rcm, Unc5h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3) (Rostral cerebellar malformation protein). | |||||
|
UNC5D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.069262 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6UXZ4, Q8WYP7 | Gene names | UNC5D, KIAA1777, UNC5H4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.069192 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
SEM3D_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.999362 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3A_HUMAN
|
||||||
| NC score | 0.994718 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3A_MOUSE
|
||||||
| NC score | 0.994398 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3E_HUMAN
|
||||||
| NC score | 0.992714 (rank : 5) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM3E_MOUSE
|
||||||
| NC score | 0.992548 (rank : 6) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3F_MOUSE
|
||||||
| NC score | 0.990777 (rank : 7) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM3C_HUMAN
|
||||||
| NC score | 0.990626 (rank : 8) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3B_MOUSE
|
||||||
| NC score | 0.990618 (rank : 9) | θ value | 1.3161e-184 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM3F_HUMAN
|
||||||
| NC score | 0.990227 (rank : 10) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM3C_MOUSE
|
||||||
| NC score | 0.990084 (rank : 11) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3B_HUMAN
|
||||||
| NC score | 0.990024 (rank : 12) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM4C_MOUSE
|
||||||
| NC score | 0.964212 (rank : 13) | θ value | 2.30293e-96 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4D_HUMAN
|
||||||
| NC score | 0.962159 (rank : 14) | θ value | 3.0147e-88 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.962044 (rank : 15) | θ value | 2.01746e-92 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.959720 (rank : 16) | θ value | 5.14231e-88 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4G_HUMAN
|
||||||
| NC score | 0.959607 (rank : 17) | θ value | 1.65422e-86 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4F_MOUSE
|
||||||
| NC score | 0.959449 (rank : 18) | θ value | 4.22625e-74 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4F_HUMAN
|
||||||
| NC score | 0.959295 (rank : 19) | θ value | 2.31364e-80 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM4C_HUMAN
|
||||||
| NC score | 0.958793 (rank : 20) | θ value | 7.42547e-87 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.955510 (rank : 21) | θ value | 3.56943e-81 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4B_MOUSE
|
||||||
| NC score | 0.953633 (rank : 22) | θ value | 7.19216e-82 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.953505 (rank : 23) | θ value | 1.18825e-76 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4A_MOUSE
|
||||||
| NC score | 0.952170 (rank : 24) | θ value | 3.34864e-71 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM7A_HUMAN
|
||||||
| NC score | 0.942386 (rank : 25) | θ value | 4.70527e-49 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.941245 (rank : 26) | θ value | 2.82823e-78 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.940918 (rank : 27) | θ value | 2.17053e-70 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.940214 (rank : 28) | θ value | 5.8972e-76 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM7A_MOUSE
|
||||||
| NC score | 0.940053 (rank : 29) | θ value | 1.85029e-45 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.939672 (rank : 30) | θ value | 2.91998e-83 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.939010 (rank : 31) | θ value | 2.16048e-86 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.934490 (rank : 32) | θ value | 1.50663e-63 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.932892 (rank : 33) | θ value | 3.71096e-62 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.926934 (rank : 34) | θ value | 4.52582e-68 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM5A_MOUSE
|
||||||
| NC score | 0.788583 (rank : 35) | θ value | 1.50663e-63 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM5B_HUMAN
|
||||||
| NC score | 0.788445 (rank : 36) | θ value | 6.11685e-65 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM5B_MOUSE
|
||||||
| NC score | 0.785423 (rank : 37) | θ value | 1.6097e-65 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5A_HUMAN
|
||||||
| NC score | 0.783895 (rank : 38) | θ value | 1.77973e-64 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.327039 (rank : 39) | θ value | 0.000461057 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXC1_MOUSE
|
||||||
| NC score | 0.313570 (rank : 40) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_HUMAN
|
||||||
| NC score | 0.308199 (rank : 41) | θ value | 0.00228821 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXA4_MOUSE
|
||||||
| NC score | 0.304137 (rank : 42) | θ value | 7.1131e-05 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.303465 (rank : 43) | θ value | 0.000602161 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA2_MOUSE
|
||||||
| NC score | 0.296638 (rank : 44) | θ value | 2.21117e-06 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA2_HUMAN
|
||||||
| NC score | 0.295367 (rank : 45) | θ value | 2.44474e-05 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.281703 (rank : 46) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA3_HUMAN
|
||||||
| NC score | 0.275198 (rank : 47) | θ value | 0.0193708 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.272333 (rank : 48) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.262715 (rank : 49) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.262238 (rank : 50) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.236935 (rank : 51) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.235954 (rank : 52) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.233415 (rank : 53) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PROP_HUMAN
|
||||||
| NC score | 0.087012 (rank : 54) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |

|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.084607 (rank : 55) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
UNC5D_HUMAN
|
||||||
| NC score | 0.069262 (rank : 56) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6UXZ4, Q8WYP7 | Gene names | UNC5D, KIAA1777, UNC5H4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5D_MOUSE
|
||||||
| NC score | 0.069192 (rank : 57) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
SPON1_MOUSE
|
||||||
| NC score | 0.064588 (rank : 58) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
SPON1_HUMAN
|
||||||
| NC score | 0.063124 (rank : 59) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
UNC5B_HUMAN
|
||||||
| NC score | 0.058621 (rank : 60) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| NC score | 0.058594 (rank : 61) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
CD79B_MOUSE
|
||||||
| NC score | 0.055610 (rank : 62) | θ value | 0.0431538 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15530 | Gene names | Cd79b, Igb | |||
|
Domain Architecture |

|
|||||
| Description | B-cell antigen receptor complex-associated protein beta-chain precursor (B-cell-specific glycoprotein B29) (Immunoglobulin- associated B29 protein) (IG-beta) (CD79b antigen). | |||||
|
UNC5A_MOUSE
|
||||||
| NC score | 0.054932 (rank : 63) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
UNC5C_MOUSE
|
||||||
| NC score | 0.054302 (rank : 64) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08747, Q8CD16 | Gene names | Unc5c, Rcm, Unc5h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3) (Rostral cerebellar malformation protein). | |||||
|
UNC5C_HUMAN
|
||||||
| NC score | 0.053354 (rank : 65) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
CILP1_MOUSE
|
||||||
| NC score | 0.041554 (rank : 66) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q66K08, Q7TSS0, Q7TSS1, Q8BV01 | Gene names | Cilp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 1 precursor (CILP-1) [Contains: Cartilage intermediate layer protein 1 C1; Cartilage intermediate layer protein 1 C2]. | |||||
|
CD79B_HUMAN
|
||||||
| NC score | 0.038121 (rank : 67) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40259, Q9BU06 | Gene names | CD79B, B29, IGB | |||
|
Domain Architecture |

|
|||||
| Description | B-cell antigen receptor complex-associated protein beta-chain precursor (B-cell-specific glycoprotein B29) (Immunoglobulin- associated B29 protein) (IG-beta) (CD79b antigen). | |||||
|
CNTN4_HUMAN
|
||||||
| NC score | 0.037893 (rank : 68) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
CNTN4_MOUSE
|
||||||
| NC score | 0.035930 (rank : 69) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q69Z26, Q8BRT6, Q8CBD3 | Gene names | Cntn4, Kiaa3024 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
CILP1_HUMAN
|
||||||
| NC score | 0.035681 (rank : 70) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75339, Q6UW99, Q8IYI5 | Gene names | CILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 1 precursor (CILP-1) (Cartilage intermediate-layer protein) [Contains: Cartilage intermediate layer protein 1 C1; Cartilage intermediate layer protein 1 C2]. | |||||
|
IRPL2_MOUSE
|
||||||
| NC score | 0.032537 (rank : 71) | θ value | 0.0330416 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ERS6, Q9ER66 | Gene names | Il1rapl2 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 2 precursor (IL1RAPL-2-related protein) (TIGIRR-1). | |||||
|
IRPL2_HUMAN
|
||||||
| NC score | 0.031601 (rank : 72) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NP60, Q9NZN0 | Gene names | IL1RAPL2, IL1R9 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 2 precursor (IL1RAPL-2-related protein) (Interleukin-1 receptor 9) (IL-1R9) (IL-1 receptor accessory protein-like 2) (Three immunoglobulin domain- containing IL-1 receptor-related 1) (TIGIRR-1). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.028682 (rank : 73) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
IRPL1_MOUSE
|
||||||
| NC score | 0.024495 (rank : 74) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59823 | Gene names | Il1rapl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 1 precursor (IL1RAPL-1). | |||||
|
IRPL1_HUMAN
|
||||||
| NC score | 0.024172 (rank : 75) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZN1, Q9UJ53 | Gene names | IL1RAPL1, OPHN4 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked interleukin-1 receptor accessory protein-like 1 precursor (IL1RAPL-1) (Oligophrenin-4) (Three immunoglobulin domain-containing IL-1 receptor-related 2) (TIGIRR-2). | |||||
|
L1CAM_HUMAN
|
||||||
| NC score | 0.021034 (rank : 76) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P32004, Q8TA87 | Gene names | L1CAM, CAML1, MIC5 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.020986 (rank : 77) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
L1CAM_MOUSE
|
||||||
| NC score | 0.019377 (rank : 78) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11627 | Gene names | L1cam, Caml1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
CNTN6_HUMAN
|
||||||
| NC score | 0.016205 (rank : 79) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
AMGO1_HUMAN
|
||||||
| NC score | 0.015487 (rank : 80) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86WK6, Q8IW71, Q9ULQ7 | Gene names | AMIGO1, ALI2, AMIGO, KIAA1163 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amphoterin-induced protein 1 precursor (AMIGO-1) (Alivin-2). | |||||
|
CNTN5_HUMAN
|
||||||
| NC score | 0.015319 (rank : 81) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94779, O94780 | Gene names | CNTN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2) (hNB-2). | |||||
|
PROX1_HUMAN
|
||||||
| NC score | 0.014718 (rank : 82) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
PROX1_MOUSE
|
||||||
| NC score | 0.014657 (rank : 83) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
KV6E_MOUSE
|
||||||
| NC score | 0.012958 (rank : 84) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01679 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region J539. | |||||
|
KV6J_MOUSE
|
||||||
| NC score | 0.012613 (rank : 85) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04944 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-VI region NQ5-78.2.6. | |||||
|
KV6I_MOUSE
|
||||||
| NC score | 0.012575 (rank : 86) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04943 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-VI region NQ6-8.3.1. | |||||
|
KV6G_MOUSE
|
||||||
| NC score | 0.012505 (rank : 87) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04941 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-VI region NQ2-48.2.2. | |||||
|
KV6F_MOUSE
|
||||||
| NC score | 0.012407 (rank : 88) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04940 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-VI region NQ2-17.4.1. | |||||
|
KV6C_MOUSE
|
||||||
| NC score | 0.011994 (rank : 89) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01677 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region TEPC 601/TEPC 191. | |||||
|
KV6B_MOUSE
|
||||||
| NC score | 0.011899 (rank : 90) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01676 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region XRPC 24. | |||||
|
KV6D_MOUSE
|
||||||
| NC score | 0.011829 (rank : 91) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01678 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region SAPC 10. | |||||
|
KV6H_MOUSE
|
||||||
| NC score | 0.011711 (rank : 92) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04942 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-VI region NQ5-61.1.2. | |||||
|
KV6A_MOUSE
|
||||||
| NC score | 0.011010 (rank : 93) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01675 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-VI region XRPC 44. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.010301 (rank : 94) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRI34_HUMAN
|
||||||
| NC score | 0.010253 (rank : 95) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYJ4, Q9C016, Q9HCR0, Q9HCR1, Q9HCR2 | Gene names | TRIM34, IFP1, RNF21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 34 (Interferon-responsive finger protein 1) (RING finger protein 21). | |||||
|
LRRN1_MOUSE
|
||||||
| NC score | 0.010100 (rank : 96) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 508 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61809, Q3USY1, Q69ZI0 | Gene names | Lrrn1, Kiaa1497 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats neuronal protein 1 precursor (Neuronal leucine- rich repeat protein 1) (NLRR-1). | |||||
|
LRRN1_HUMAN
|
||||||
| NC score | 0.009388 (rank : 97) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UXK5, Q3LID5, Q8IYV5, Q9H8V1, Q9P231 | Gene names | LRRN1, KIAA1497 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats neuronal protein 1 precursor (Neuronal leucine- rich repeat protein 1) (NLRR-1). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.008221 (rank : 98) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CEAM6_HUMAN
|
||||||
| NC score | 0.007801 (rank : 99) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P40199, Q14920 | Gene names | CEACAM6, NCA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 6 precursor (Normal cross-reacting antigen) (Nonspecific crossreacting antigen) (CD66c antigen). | |||||
|
KV1W_HUMAN
|
||||||
| NC score | 0.007466 (rank : 100) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04431 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-I region Walker precursor. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.006803 (rank : 101) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
KV2B_HUMAN
|
||||||
| NC score | 0.006204 (rank : 102) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P01615 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region FR. | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.004861 (rank : 103) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
TMC1_HUMAN
|
||||||
| NC score | 0.002549 (rank : 104) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI8 | Gene names | TMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1. | |||||
|
ITB3_MOUSE
|
||||||
| NC score | 0.001081 (rank : 105) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.000488 (rank : 106) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.000464 (rank : 107) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | -0.001014 (rank : 108) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MERTK_HUMAN
|
||||||
| NC score | -0.001820 (rank : 109) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12866, Q9HBB4 | Gene names | MERTK, MER | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase MER precursor (EC 2.7.10.1) (C- mer) (Receptor tyrosine kinase MerTK). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | -0.001920 (rank : 110) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||