Please be patient as the page loads
|
MCES_MOUSE
|
||||||
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCES_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.915531 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
MCES_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 3) | NC score | 0.036331 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 4) | NC score | 0.082149 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.080283 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.079743 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
WDR81_HUMAN
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.048435 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q562E7, Q24JP6, Q8N277, Q8N3F3 | Gene names | WDR81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.083156 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.037203 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.047941 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.062860 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.031726 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.037214 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.054280 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LYAR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.083010 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.034032 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
KIF3B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.036198 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
SEC62_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.096148 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99442, O00682, O00729 | Gene names | TLOC1, SEC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1). | |||||
|
CD2B2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.048891 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95400, Q9ULP2 | Gene names | CD2BP2, KIAA1178 | |||
|
Domain Architecture |

|
|||||
| Description | CD2 antigen cytoplasmic tail-binding protein 2 (CD2 cytoplasmic domain-binding protein) (CD2 tail-binding protein). | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.036113 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.073275 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.026620 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
UT14A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.038392 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.053424 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TRI37_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.027794 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.033506 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.030405 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DEK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.067925 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.029382 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.034801 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MIPO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.045990 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TD10, Q7Z3J0, Q8IV14 | Gene names | MIPOL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mirror-image polydactyly gene 1 protein. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.034822 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SEC62_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.073839 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.029417 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.017057 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
SYEP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.040151 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
TCAL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.031378 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.027694 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.033082 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NOL10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.038018 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.029120 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.010876 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.054017 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.023068 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.039878 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.012931 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.024434 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.020562 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.029166 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
ROBO2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.007538 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.028162 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.027995 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.014209 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
BORG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.022210 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKI2, O95353, Q9UQJ0 | Gene names | CDC42EP3, BORG2, CEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein). | |||||
|
CN140_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.032045 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.021894 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.025393 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.031027 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
KIF3C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.031440 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.036363 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.011169 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.011092 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.013827 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CAYP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.021690 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.004655 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CFDP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.025771 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88271, O70565, Q9JMA5 | Gene names | Cfdp1, Bcnt, Cfdp, Cp27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur) (27 kDa craniofacial protein) (Protein Cp27). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.046594 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.044323 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.022066 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
NOL10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.031277 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5RJG1 | Gene names | Nol10, Gm67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.017134 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.047315 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RN169_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.023688 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
TNNT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.022414 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
ARRS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.009649 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |
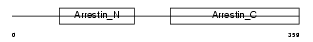
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
BAP31_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.019501 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51572, Q13836, Q96CF0 | Gene names | BCAP31, BAP31, DXS1357E | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31) (Protein CDM) (6C6-AG tumor-associated antigen). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.017159 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.031011 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CN037_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.015569 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.026402 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
HDAC9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.014873 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.019983 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.027352 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.021817 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.006378 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
STX18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.022684 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDS8, Q3TDJ0, Q9D8T6 | Gene names | Stx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18. | |||||
|
TRI14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.007084 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVW3, Q6A0C3, Q762I6, Q9D3G8 | Gene names | Trim14, Kiaa0129, Pub | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 14 (PU.1-binding protein). | |||||
|
ZCHC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.016269 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.067266 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.058886 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053144 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055926 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.054538 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
MCES_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
MCES_HUMAN
|
||||||
| NC score | 0.915531 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
SEC62_HUMAN
|
||||||
| NC score | 0.096148 (rank : 3) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99442, O00682, O00729 | Gene names | TLOC1, SEC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.083156 (rank : 4) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
LYAR_HUMAN
|
||||||
| NC score | 0.083010 (rank : 5) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.082149 (rank : 6) | θ value | 0.0431538 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.080283 (rank : 7) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.079743 (rank : 8) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SEC62_MOUSE
|
||||||
| NC score | 0.073839 (rank : 9) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.073275 (rank : 10) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.067925 (rank : 11) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.067266 (rank : 12) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.062860 (rank : 13) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.058886 (rank : 14) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.055926 (rank : 15) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.054538 (rank : 16) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.054280 (rank : 17) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.054017 (rank : 18) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.053424 (rank : 19) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.053144 (rank : 20) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
CD2B2_HUMAN
|
||||||
| NC score | 0.048891 (rank : 21) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95400, Q9ULP2 | Gene names | CD2BP2, KIAA1178 | |||
|
Domain Architecture |

|
|||||
| Description | CD2 antigen cytoplasmic tail-binding protein 2 (CD2 cytoplasmic domain-binding protein) (CD2 tail-binding protein). | |||||
|
WDR81_HUMAN
|
||||||
| NC score | 0.048435 (rank : 22) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q562E7, Q24JP6, Q8N277, Q8N3F3 | Gene names | WDR81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.047941 (rank : 23) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.047315 (rank : 24) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.046594 (rank : 25) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
MIPO1_HUMAN
|
||||||
| NC score | 0.045990 (rank : 26) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TD10, Q7Z3J0, Q8IV14 | Gene names | MIPOL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mirror-image polydactyly gene 1 protein. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.044323 (rank : 27) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SYEP_HUMAN
|
||||||
| NC score | 0.040151 (rank : 28) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.039878 (rank : 29) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
UT14A_HUMAN
|
||||||
| NC score | 0.038392 (rank : 30) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
NOL10_HUMAN
|
||||||
| NC score | 0.038018 (rank : 31) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.037214 (rank : 32) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.037203 (rank : 33) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.036363 (rank : 34) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.036331 (rank : 35) | θ value | 0.0148317 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
KIF3B_HUMAN
|
||||||
| NC score | 0.036198 (rank : 36) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.036113 (rank : 37) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.034822 (rank : 38) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.034801 (rank : 39) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.034032 (rank : 40) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.033506 (rank : 41) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.033082 (rank : 42) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CN140_MOUSE
|
||||||
| NC score | 0.032045 (rank : 43) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.031726 (rank : 44) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
KIF3C_MOUSE
|
||||||
| NC score | 0.031440 (rank : 45) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
TCAL4_HUMAN
|
||||||
| NC score | 0.031378 (rank : 46) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
NOL10_MOUSE
|
||||||
| NC score | 0.031277 (rank : 47) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5RJG1 | Gene names | Nol10, Gm67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.031027 (rank : 48) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.031011 (rank : 49) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.030405 (rank : 50) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.029417 (rank : 51) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.029382 (rank : 52) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.029166 (rank : 53) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.029120 (rank : 54) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.028162 (rank : 55) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.027995 (rank : 56) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TRI37_MOUSE
|
||||||
| NC score | 0.027794 (rank : 57) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.027694 (rank : 58) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.027352 (rank : 59) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.026620 (rank : 60) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.026402 (rank : 61) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
CFDP1_MOUSE
|
||||||
| NC score | 0.025771 (rank : 62) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88271, O70565, Q9JMA5 | Gene names | Cfdp1, Bcnt, Cfdp, Cp27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur) (27 kDa craniofacial protein) (Protein Cp27). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.025393 (rank : 63) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.024434 (rank : 64) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.023688 (rank : 65) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.023068 (rank : 66) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
STX18_MOUSE
|
||||||
| NC score | 0.022684 (rank : 67) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDS8, Q3TDJ0, Q9D8T6 | Gene names | Stx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18. | |||||
|
TNNT3_MOUSE
|
||||||
| NC score | 0.022414 (rank : 68) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
BORG2_HUMAN
|
||||||
| NC score | 0.022210 (rank : 69) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKI2, O95353, Q9UQJ0 | Gene names | CDC42EP3, BORG2, CEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.022066 (rank : 70) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.021894 (rank : 71) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.021817 (rank : 72) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.021690 (rank : 73) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.020562 (rank : 74) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.019983 (rank : 75) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
BAP31_HUMAN
|
||||||
| NC score | 0.019501 (rank : 76) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51572, Q13836, Q96CF0 | Gene names | BCAP31, BAP31, DXS1357E | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31) (Protein CDM) (6C6-AG tumor-associated antigen). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.017159 (rank : 77) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.017134 (rank : 78) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.017057 (rank : 79) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
ZCHC8_HUMAN
|
||||||
| NC score | 0.016269 (rank : 80) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.015569 (rank : 81) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
HDAC9_MOUSE
|
||||||
| NC score | 0.014873 (rank : 82) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N13, Q9EPT2 | Gene names | Hdac9, Hdac7b, Hdrp, Mitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.014209 (rank : 83) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.013827 (rank : 84) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.012931 (rank : 85) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.011169 (rank : 86) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.011092 (rank : 87) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.010876 (rank : 88) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
ARRS_HUMAN
|
||||||
| NC score | 0.009649 (rank : 89) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
ROBO2_HUMAN
|
||||||
| NC score | 0.007538 (rank : 90) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
TRI14_MOUSE
|
||||||
| NC score | 0.007084 (rank : 91) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVW3, Q6A0C3, Q762I6, Q9D3G8 | Gene names | Trim14, Kiaa0129, Pub | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 14 (PU.1-binding protein). | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.006378 (rank : 92) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.004655 (rank : 93) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||