Please be patient as the page loads
|
ITIH4_HUMAN
|
||||||
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITIH4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 1.36511e-173 (rank : 2) | NC score | 0.971576 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 5.73535e-172 (rank : 3) | NC score | 0.971223 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 2.97304e-136 (rank : 4) | NC score | 0.952003 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 2.13063e-134 (rank : 5) | NC score | 0.963252 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 6.19918e-134 (rank : 6) | NC score | 0.963784 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 7.57797e-132 (rank : 7) | NC score | 0.955242 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 8) | NC score | 0.344964 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 9) | NC score | 0.375774 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 10) | NC score | 0.352299 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 11) | NC score | 0.311470 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 12) | NC score | 0.311689 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
ANTR2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 13) | NC score | 0.088993 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6DFX2, Q99L17 | Gene names | Antxr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anthrax toxin receptor 2 precursor. | |||||
|
ANTR2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 14) | NC score | 0.079766 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58335, Q59E98, Q5JPE9, Q86UI1, Q8N4J8, Q8NB13, Q96NC7 | Gene names | ANTXR2, CMG2 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 2 precursor (Capillary morphogenesis gene 2 protein) (CMG-2). | |||||
|
CO6A1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.034466 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
CO6A1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.039670 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q04857 | Gene names | Col6a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
MATN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.059169 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
ITAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.029940 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
FCGR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.016805 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26151 | Gene names | Fcgr1, Fcg1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.033362 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.016477 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
CAYP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.022343 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.016121 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.013456 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.027599 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
MATN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.052438 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |
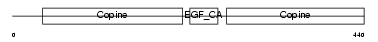
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
EM55_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.011454 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |
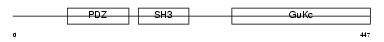
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
MATN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.020906 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
PABP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.009654 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.009633 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.014191 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
EM55_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.010304 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |
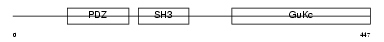
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
MATN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.025712 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
POLS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.012466 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.014536 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CN155_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.011310 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
K1718_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.007495 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.011367 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
SUSD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.009028 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.007305 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.006441 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.001615 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.010299 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.005972 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
ITAX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.021217 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
SPHK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.008853 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |
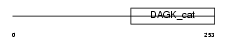
|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.971576 (rank : 2) | θ value | 1.36511e-173 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.971223 (rank : 3) | θ value | 5.73535e-172 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.963784 (rank : 4) | θ value | 6.19918e-134 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.963252 (rank : 5) | θ value | 2.13063e-134 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.955242 (rank : 6) | θ value | 7.57797e-132 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.952003 (rank : 7) | θ value | 2.97304e-136 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.375774 (rank : 8) | θ value | 1.47974e-10 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.352299 (rank : 9) | θ value | 8.11959e-09 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.344964 (rank : 10) | θ value | 5.08577e-11 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.311689 (rank : 11) | θ value | 0.000270298 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.311470 (rank : 12) | θ value | 7.1131e-05 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
ANTR2_MOUSE
|
||||||
| NC score | 0.088993 (rank : 13) | θ value | 0.000461057 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6DFX2, Q99L17 | Gene names | Antxr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anthrax toxin receptor 2 precursor. | |||||
|
ANTR2_HUMAN
|
||||||
| NC score | 0.079766 (rank : 14) | θ value | 0.00665767 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58335, Q59E98, Q5JPE9, Q86UI1, Q8N4J8, Q8NB13, Q96NC7 | Gene names | ANTXR2, CMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 2 precursor (Capillary morphogenesis gene 2 protein) (CMG-2). | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.059169 (rank : 15) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_HUMAN
|
||||||
| NC score | 0.052438 (rank : 16) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
CO6A1_MOUSE
|
||||||
| NC score | 0.039670 (rank : 17) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q04857 | Gene names | Col6a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
CO6A1_HUMAN
|
||||||
| NC score | 0.034466 (rank : 18) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.033362 (rank : 19) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ITAM_MOUSE
|
||||||
| NC score | 0.029940 (rank : 20) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.027599 (rank : 21) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.025712 (rank : 22) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.022343 (rank : 23) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
ITAX_HUMAN
|
||||||
| NC score | 0.021217 (rank : 24) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.020906 (rank : 25) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
FCGR1_MOUSE
|
||||||
| NC score | 0.016805 (rank : 26) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26151 | Gene names | Fcgr1, Fcg1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.016477 (rank : 27) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.016121 (rank : 28) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.014536 (rank : 29) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.014191 (rank : 30) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.013456 (rank : 31) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
POLS_MOUSE
|
||||||
| NC score | 0.012466 (rank : 32) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
EM55_MOUSE
|
||||||
| NC score | 0.011454 (rank : 33) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.011367 (rank : 34) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.011310 (rank : 35) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
EM55_HUMAN
|
||||||
| NC score | 0.010304 (rank : 36) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.010299 (rank : 37) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
PABP1_HUMAN
|
||||||
| NC score | 0.009654 (rank : 38) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| NC score | 0.009633 (rank : 39) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
SUSD4_HUMAN
|
||||||
| NC score | 0.009028 (rank : 40) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SPHK2_HUMAN
|
||||||
| NC score | 0.008853 (rank : 41) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
K1718_MOUSE
|
||||||
| NC score | 0.007495 (rank : 42) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.007305 (rank : 43) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.006441 (rank : 44) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.005972 (rank : 45) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | 0.001615 (rank : 46) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||