Please be patient as the page loads
|
LHR2A_HUMAN
|
||||||
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LHR2A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989424 (rank : 2) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
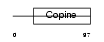
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 3) | NC score | 0.630753 (rank : 3) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 4) | NC score | 0.344635 (rank : 5) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 5) | NC score | 0.375774 (rank : 4) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 6) | NC score | 0.312691 (rank : 6) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.306928 (rank : 7) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 8) | NC score | 0.299780 (rank : 8) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.028585 (rank : 20) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
CASC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.034526 (rank : 18) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
PTHB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.034107 (rank : 19) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q811G0, P97498, Q3UMR2, Q7TQH9, Q8BKD7, Q8K0T5 | Gene names | Pthb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTHB1 (Parathyroid hormone-responsive B1 gene protein homolog). | |||||
|
EGR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.004116 (rank : 24) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06889 | Gene names | EGR3, PILOT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 3 (EGR-3) (Zinc finger protein pilot). | |||||
|
EGR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.003535 (rank : 25) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43300, Q9R276 | Gene names | Egr3, Egr-3 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 3 (EGR-3). | |||||
|
MDR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.005007 (rank : 23) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08183, Q12755, Q14812 | Gene names | ABCB1, MDR1, PGY1 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 1 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1) (P-glycoprotein 1) (CD243 antigen). | |||||
|
AYT1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.010791 (rank : 21) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.010427 (rank : 22) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.055041 (rank : 12) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.054753 (rank : 13) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.205624 (rank : 10) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.233689 (rank : 9) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
PARP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.053460 (rank : 14) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.051535 (rank : 15) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.055724 (rank : 11) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
PARP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.050838 (rank : 16) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.050013 (rank : 17) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.989424 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.630753 (rank : 3) | θ value | 5.76516e-39 (rank : 3) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.375774 (rank : 4) | θ value | 1.47974e-10 (rank : 5) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.344635 (rank : 5) | θ value | 3.89403e-11 (rank : 4) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.312691 (rank : 6) | θ value | 1.99992e-07 (rank : 6) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.306928 (rank : 7) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.299780 (rank : 8) | θ value | 9.29e-05 (rank : 8) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.233689 (rank : 9) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.205624 (rank : 10) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
PARP2_HUMAN
|
||||||
| NC score | 0.055724 (rank : 11) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UGN5, Q8TEU4, Q9NUV2, Q9UMR4, Q9Y6C8 | Gene names | PARP2, ADPRT2, ADPRTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (hPARP-2). | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.055041 (rank : 12) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.054753 (rank : 13) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP1_HUMAN
|
||||||
| NC score | 0.053460 (rank : 14) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P09874, Q8IUZ9 | Gene names | PARP1, ADPRT, PPOL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1). | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.051535 (rank : 15) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
PARP2_MOUSE
|
||||||
| NC score | 0.050838 (rank : 16) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O88554, Q99N29 | Gene names | Parp2, Adprt2, Adprtl2, Aspartl2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 2 (EC 2.4.2.30) (PARP-2) (NAD(+) ADP- ribosyltransferase 2) (Poly[ADP-ribose] synthetase 2) (pADPRT-2) (mPARP-2). | |||||
|
PARP3_HUMAN
|
||||||
| NC score | 0.050013 (rank : 17) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
CASC5_MOUSE
|
||||||
| NC score | 0.034526 (rank : 18) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
PTHB1_MOUSE
|
||||||
| NC score | 0.034107 (rank : 19) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q811G0, P97498, Q3UMR2, Q7TQH9, Q8BKD7, Q8K0T5 | Gene names | Pthb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTHB1 (Parathyroid hormone-responsive B1 gene protein homolog). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.028585 (rank : 20) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
AYT1B_MOUSE
|
||||||
| NC score | 0.010791 (rank : 21) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| NC score | 0.010427 (rank : 22) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
MDR1_HUMAN
|
||||||
| NC score | 0.005007 (rank : 23) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08183, Q12755, Q14812 | Gene names | ABCB1, MDR1, PGY1 | |||
|
Domain Architecture |

|
|||||
| Description | Multidrug resistance protein 1 (EC 3.6.3.44) (ATP-binding cassette sub-family B member 1) (P-glycoprotein 1) (CD243 antigen). | |||||
|
EGR3_HUMAN
|
||||||
| NC score | 0.004116 (rank : 24) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06889 | Gene names | EGR3, PILOT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 3 (EGR-3) (Zinc finger protein pilot). | |||||
|
EGR3_MOUSE
|
||||||
| NC score | 0.003535 (rank : 25) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 16 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43300, Q9R276 | Gene names | Egr3, Egr-3 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 3 (EGR-3). | |||||