Please be patient as the page loads
|
ITIH2_HUMAN
|
||||||
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITIH2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995163 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.971754 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.972040 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 1.04281e-181 (rank : 5) | NC score | 0.969348 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH1_HUMAN
|
||||||
| θ value | 2.48782e-175 (rank : 6) | NC score | 0.965036 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 2.97304e-136 (rank : 7) | NC score | 0.952003 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 8) | NC score | 0.304280 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 9) | NC score | 0.305760 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.227211 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
PTEN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.027486 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |
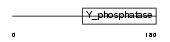
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
PTEN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.027484 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |
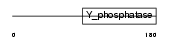
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
FA29A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.027993 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.012317 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.008963 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
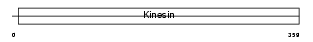
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
KIF3C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.008929 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |
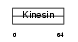
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.004205 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.015065 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.003954 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.007505 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.009397 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.008565 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
UGPA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.014657 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16851, Q86Y81, Q9BU15 | Gene names | UGP2 | |||
|
Domain Architecture |
|
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.007039 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.003883 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.009114 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
FANCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.008637 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
|
PLK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | -0.001683 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64702, Q6PEP6, Q78EG6, Q8R0I5, Q9CVR6, Q9CVU6 | Gene names | Plk4, Sak, Stk18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
UGPA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.012714 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZJ5, Q8R3D2 | Gene names | Ugp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.205624 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.189843 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.995163 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.972040 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.971754 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.969348 (rank : 5) | θ value | 1.04281e-181 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.965036 (rank : 6) | θ value | 2.48782e-175 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.952003 (rank : 7) | θ value | 2.97304e-136 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.305760 (rank : 8) | θ value | 0.000158464 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.304280 (rank : 9) | θ value | 0.000158464 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.227211 (rank : 10) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.205624 (rank : 11) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.189843 (rank : 12) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
FA29A_HUMAN
|
||||||
| NC score | 0.027993 (rank : 13) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
PTEN_HUMAN
|
||||||
| NC score | 0.027486 (rank : 14) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
PTEN_MOUSE
|
||||||
| NC score | 0.027484 (rank : 15) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.015065 (rank : 16) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
UGPA2_HUMAN
|
||||||
| NC score | 0.014657 (rank : 17) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16851, Q86Y81, Q9BU15 | Gene names | UGP2 | |||
|
Domain Architecture |

|
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
UGPA2_MOUSE
|
||||||
| NC score | 0.012714 (rank : 18) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZJ5, Q8R3D2 | Gene names | Ugp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.012317 (rank : 19) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.009397 (rank : 20) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.009114 (rank : 21) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.008963 (rank : 22) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
KIF3C_MOUSE
|
||||||
| NC score | 0.008929 (rank : 23) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35066, O35229 | Gene names | Kif3c | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
FANCA_HUMAN
|
||||||
| NC score | 0.008637 (rank : 24) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.008565 (rank : 25) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.007505 (rank : 26) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.007039 (rank : 27) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.004205 (rank : 28) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.003954 (rank : 29) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.003883 (rank : 30) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
PLK4_MOUSE
|
||||||
| NC score | -0.001683 (rank : 31) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64702, Q6PEP6, Q78EG6, Q8R0I5, Q9CVR6, Q9CVU6 | Gene names | Plk4, Sak, Stk18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||