Please be patient as the page loads
|
ITIH1_MOUSE
|
||||||
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITIH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997070 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987534 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987059 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 1.04281e-181 (rank : 5) | NC score | 0.969348 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 3.47146e-177 (rank : 6) | NC score | 0.969596 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 2.13063e-134 (rank : 7) | NC score | 0.963252 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 8) | NC score | 0.312691 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 9) | NC score | 0.306030 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 10) | NC score | 0.275438 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 11) | NC score | 0.302912 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 12) | NC score | 0.279534 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
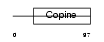
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
MATN2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.037152 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.034714 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.060949 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
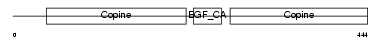
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.053131 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |
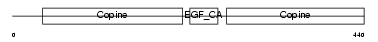
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.031924 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.012077 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.002202 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
WDR81_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.013855 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5ND34, Q3TA90, Q3TUS9, Q6KAR1 | Gene names | Wdr81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
ACPH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.011597 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13798, Q9BQ33, Q9P0Y2 | Gene names | APEH | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase) (Oxidized protein hydrolase) (OPH) (DNF15S2 protein). | |||||
|
CB013_MOUSE
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.014178 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D842, Q8BZL5, Q99LX6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13 homolog. | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.026943 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
ITAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.023387 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
DPYL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.009002 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62188 | Gene names | Dpysl3, Drp3, Ulip | |||
|
Domain Architecture |
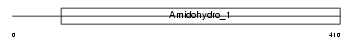
|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein). | |||||
|
MET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | -0.000942 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08581, O60366, Q12875, Q9UDX7, Q9UPL8 | Gene names | MET | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.006501 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.002457 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.002833 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.000515 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
WDR81_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.011839 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q562E7, Q24JP6, Q8N277, Q8N3F3 | Gene names | WDR81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.997070 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.987534 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.987059 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.969596 (rank : 5) | θ value | 3.47146e-177 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.969348 (rank : 6) | θ value | 1.04281e-181 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.963252 (rank : 7) | θ value | 2.13063e-134 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.312691 (rank : 8) | θ value | 1.99992e-07 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.306030 (rank : 9) | θ value | 0.00020696 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.302912 (rank : 10) | θ value | 0.000602161 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.279534 (rank : 11) | θ value | 0.00509761 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.275438 (rank : 12) | θ value | 0.000270298 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.060949 (rank : 13) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_HUMAN
|
||||||
| NC score | 0.053131 (rank : 14) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN2_MOUSE
|
||||||
| NC score | 0.037152 (rank : 15) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.034714 (rank : 16) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.031924 (rank : 17) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.026943 (rank : 18) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
ITAM_MOUSE
|
||||||
| NC score | 0.023387 (rank : 19) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
CB013_MOUSE
|
||||||
| NC score | 0.014178 (rank : 20) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D842, Q8BZL5, Q99LX6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13 homolog. | |||||
|
WDR81_MOUSE
|
||||||
| NC score | 0.013855 (rank : 21) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5ND34, Q3TA90, Q3TUS9, Q6KAR1 | Gene names | Wdr81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.012077 (rank : 22) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
WDR81_HUMAN
|
||||||
| NC score | 0.011839 (rank : 23) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q562E7, Q24JP6, Q8N277, Q8N3F3 | Gene names | WDR81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
ACPH_HUMAN
|
||||||
| NC score | 0.011597 (rank : 24) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13798, Q9BQ33, Q9P0Y2 | Gene names | APEH | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase) (Oxidized protein hydrolase) (OPH) (DNF15S2 protein). | |||||
|
DPYL3_MOUSE
|
||||||
| NC score | 0.009002 (rank : 25) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62188 | Gene names | Dpysl3, Drp3, Ulip | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.006501 (rank : 26) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.002833 (rank : 27) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.002457 (rank : 28) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.002202 (rank : 29) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.000515 (rank : 30) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
MET_HUMAN
|
||||||
| NC score | -0.000942 (rank : 31) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08581, O60366, Q12875, Q9UDX7, Q9UPL8 | Gene names | MET | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||