Please be patient as the page loads
|
ITIH1_HUMAN
|
||||||
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITIH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997070 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.986979 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.986326 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 2.48782e-175 (rank : 5) | NC score | 0.965036 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 3.71755e-171 (rank : 6) | NC score | 0.965392 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 6.19918e-134 (rank : 7) | NC score | 0.963784 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 8) | NC score | 0.344635 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.311930 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
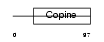
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 10) | NC score | 0.307037 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.304414 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.286447 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
MATN2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.029136 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
DTBP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.034348 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |
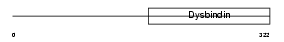
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.005808 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MATN2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.026879 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.049197 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
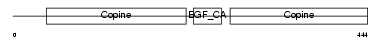
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
CA026_MOUSE
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.018093 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
DPYL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.012734 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62188 | Gene names | Dpysl3, Drp3, Ulip | |||
|
Domain Architecture |
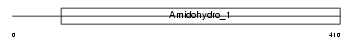
|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein). | |||||
|
TLK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.002502 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.006192 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
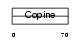
MATN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.020152 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
TLK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.002129 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
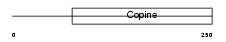
MATN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.019884 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
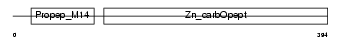
CBPB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.005084 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15086, O60834, Q96BQ8 | Gene names | CPB1, CPB, PCPB | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase B precursor (EC 3.4.17.2) (Pancreas-specific protein) (PASP). | |||||
|
MATN4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.023433 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
UBQL4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.006363 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
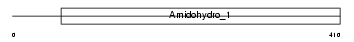
DPYL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.007746 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14195, Q93012 | Gene names | DPYSL3, CRMP4, DRP3, ULIP | |||
|
Domain Architecture |
|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein) (Collapsin response mediator protein 4) (CRMP-4). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.006353 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.007664 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.997070 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.986979 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.986326 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.965392 (rank : 5) | θ value | 3.71755e-171 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.965036 (rank : 6) | θ value | 2.48782e-175 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.963784 (rank : 7) | θ value | 6.19918e-134 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.344635 (rank : 8) | θ value | 3.89403e-11 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.311930 (rank : 9) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.307037 (rank : 10) | θ value | 0.000121331 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.304414 (rank : 11) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.286447 (rank : 12) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.049197 (rank : 13) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
DTBP1_MOUSE
|
||||||
| NC score | 0.034348 (rank : 14) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WZ8, Q80ZN4 | Gene names | Dtnbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein homolog). | |||||
|
MATN2_MOUSE
|
||||||
| NC score | 0.029136 (rank : 15) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.026879 (rank : 16) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.023433 (rank : 17) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
MATN3_HUMAN
|
||||||
| NC score | 0.020152 (rank : 18) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
MATN3_MOUSE
|
||||||
| NC score | 0.019884 (rank : 19) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.018093 (rank : 20) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
DPYL3_MOUSE
|
||||||
| NC score | 0.012734 (rank : 21) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62188 | Gene names | Dpysl3, Drp3, Ulip | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein). | |||||
|
DPYL3_HUMAN
|
||||||
| NC score | 0.007746 (rank : 22) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14195, Q93012 | Gene names | DPYSL3, CRMP4, DRP3, ULIP | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 3 (DRP-3) (Unc-33-like phosphoprotein) (ULIP protein) (Collapsin response mediator protein 4) (CRMP-4). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.007664 (rank : 23) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
UBQL4_MOUSE
|
||||||
| NC score | 0.006363 (rank : 24) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.006353 (rank : 25) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.006192 (rank : 26) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.005808 (rank : 27) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CBPB1_HUMAN
|
||||||
| NC score | 0.005084 (rank : 28) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15086, O60834, Q96BQ8 | Gene names | CPB1, CPB, PCPB | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase B precursor (EC 3.4.17.2) (Pancreas-specific protein) (PASP). | |||||
|
TLK1_HUMAN
|
||||||
| NC score | 0.002502 (rank : 29) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
TLK1_MOUSE
|
||||||
| NC score | 0.002129 (rank : 30) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||