Please be patient as the page loads
|
ITIH3_HUMAN
|
||||||
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITIH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986979 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987534 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.971754 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.970738 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
ITIH3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.996901 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 1.36511e-173 (rank : 7) | NC score | 0.971576 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
CAC2D_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 8) | NC score | 0.349666 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 9) | NC score | 0.350310 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 10) | NC score | 0.287804 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 11) | NC score | 0.306928 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
LHR2A_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.279594 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |
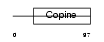
|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.020161 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.005278 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
ZN746_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.002946 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U133 | Gene names | Znf746, Zfp746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
INP4B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.016673 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1Y8 | Gene names | Inpp4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
GALA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.028839 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47212 | Gene names | Gal, Galn | |||
|
Domain Architecture |

|
|||||
| Description | Galanin precursor [Contains: Galanin; Galanin message-associated peptide (GMAP)]. | |||||
|
KPCD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.000559 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05655, Q15144 | Gene names | PRKCD | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C delta type (EC 2.7.11.13) (nPKC-delta). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.002791 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CO6A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.007037 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02788, Q05505 | Gene names | Col6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.005970 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ITAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.020996 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.014268 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.012133 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
ANKS6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.008708 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.001741 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
NSF1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.017468 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNZ2, Q7Z533, Q9H102, Q9NVL9, Q9UI06 | Gene names | NSFL1C | |||
|
Domain Architecture |
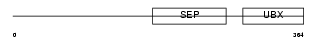
|
|||||
| Description | NSFL1 cofactor p47 (p97 cofactor p47). | |||||
|
NSF1C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.017514 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZ44, Q80Y15, Q8BYL3 | Gene names | Nsfl1c | |||
|
Domain Architecture |
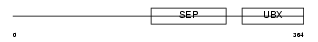
|
|||||
| Description | NSFL1 cofactor p47 (p97 cofactor p47). | |||||
|
PGFRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | -0.000052 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16234, Q96KZ7 | Gene names | PDGFRA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
I2C1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.007040 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL18, Q5TA57, Q6P4S0 | Gene names | EIF2C1, AGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Putative RNA-binding protein Q99). | |||||
|
I2C1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.007059 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJG1 | Gene names | Eif2c1, Ago1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Piwi/argonaute family protein meIF2C1). | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.002111 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.004421 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
ZN746_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.000860 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NUN9 | Gene names | ZNF746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
ABCF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.004152 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.007097 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
CCD38_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.005580 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
CO2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.007705 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
ITIH3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q06033, Q99085 | Gene names | ITIH3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH3_MOUSE
|
||||||
| NC score | 0.996901 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61704 | Gene names | Itih3 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H3 precursor (ITI heavy chain H3) (Inter-alpha-inhibitor heavy chain 3). | |||||
|
ITIH1_MOUSE
|
||||||
| NC score | 0.987534 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61702 | Gene names | Itih1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1). | |||||
|
ITIH1_HUMAN
|
||||||
| NC score | 0.986979 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19827, P78455, Q01746 | Gene names | ITIH1, IGHEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H1 precursor (ITI heavy chain H1) (Inter-alpha-inhibitor heavy chain 1) (Inter-alpha-trypsin inhibitor complex component III) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.971754 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.971576 (rank : 6) | θ value | 1.36511e-173 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.970738 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
CAC2D_MOUSE
|
||||||
| NC score | 0.350310 (rank : 8) | θ value | 4.76016e-09 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08532, O08533, O08534, O08535, O08536 | Gene names | Cacna2d1, Cacna2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
CAC2D_HUMAN
|
||||||
| NC score | 0.349666 (rank : 9) | θ value | 4.76016e-09 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54289 | Gene names | CACNA2D1, CACNL2A, CCHL2A | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyridine-sensitive L-type calcium channel subunits alpha- 2/delta precursor [Contains: L-type calcium channel subunit alpha-2; L-type calcium channel subunit delta]. | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.306928 (rank : 10) | θ value | 1.87187e-05 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.287804 (rank : 11) | θ value | 2.21117e-06 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
LHR2A_MOUSE
|
||||||
| NC score | 0.279594 (rank : 12) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99KC8, Q3TTU2, Q6UN22, Q8BHA8, Q9CTV9 | Gene names | Loh11cr2a | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein homolog. | |||||
|
GALA_MOUSE
|
||||||
| NC score | 0.028839 (rank : 13) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47212 | Gene names | Gal, Galn | |||
|
Domain Architecture |

|
|||||
| Description | Galanin precursor [Contains: Galanin; Galanin message-associated peptide (GMAP)]. | |||||
|
ITAM_MOUSE
|
||||||
| NC score | 0.020996 (rank : 14) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.020161 (rank : 15) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
NSF1C_MOUSE
|
||||||
| NC score | 0.017514 (rank : 16) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CZ44, Q80Y15, Q8BYL3 | Gene names | Nsfl1c | |||
|
Domain Architecture |

|
|||||
| Description | NSFL1 cofactor p47 (p97 cofactor p47). | |||||
|
NSF1C_HUMAN
|
||||||
| NC score | 0.017468 (rank : 17) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNZ2, Q7Z533, Q9H102, Q9NVL9, Q9UI06 | Gene names | NSFL1C | |||
|
Domain Architecture |

|
|||||
| Description | NSFL1 cofactor p47 (p97 cofactor p47). | |||||
|
INP4B_MOUSE
|
||||||
| NC score | 0.016673 (rank : 18) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1Y8 | Gene names | Inpp4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.014268 (rank : 19) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.012133 (rank : 20) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
ANKS6_HUMAN
|
||||||
| NC score | 0.008708 (rank : 21) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
CO2_MOUSE
|
||||||
| NC score | 0.007705 (rank : 22) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.007097 (rank : 23) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
I2C1_MOUSE
|
||||||
| NC score | 0.007059 (rank : 24) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJG1 | Gene names | Eif2c1, Ago1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Piwi/argonaute family protein meIF2C1). | |||||
|
I2C1_HUMAN
|
||||||
| NC score | 0.007040 (rank : 25) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL18, Q5TA57, Q6P4S0 | Gene names | EIF2C1, AGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Putative RNA-binding protein Q99). | |||||
|
CO6A2_MOUSE
|
||||||
| NC score | 0.007037 (rank : 26) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02788, Q05505 | Gene names | Col6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.005970 (rank : 27) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
CCD38_HUMAN
|
||||||
| NC score | 0.005580 (rank : 28) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.005278 (rank : 29) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.004421 (rank : 30) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
ABCF1_MOUSE
|
||||||
| NC score | 0.004152 (rank : 31) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
ZN746_MOUSE
|
||||||
| NC score | 0.002946 (rank : 32) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U133 | Gene names | Znf746, Zfp746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.002791 (rank : 33) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.002111 (rank : 34) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.001741 (rank : 35) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
ZN746_HUMAN
|
||||||
| NC score | 0.000860 (rank : 36) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NUN9 | Gene names | ZNF746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
KPCD_HUMAN
|
||||||
| NC score | 0.000559 (rank : 37) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05655, Q15144 | Gene names | PRKCD | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C delta type (EC 2.7.11.13) (nPKC-delta). | |||||
|
PGFRA_HUMAN
|
||||||
| NC score | -0.000052 (rank : 38) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16234, Q96KZ7 | Gene names | PDGFRA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||