Please be patient as the page loads
|
SPHK2_HUMAN
|
||||||
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPHK2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
SPHK2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978938 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
SPHK1_HUMAN
|
||||||
| θ value | 3.974e-56 (rank : 3) | NC score | 0.903900 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYA1, Q9HD92, Q9NY70, Q9NYL3 | Gene names | SPHK1, SPHK, SPK | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosine kinase 1 (EC 2.7.1.-) (SK 1) (SPK 1). | |||||
|
CERK1_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 4) | NC score | 0.655657 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TCT0, Q9BYB3, Q9UGE5 | Gene names | CERK, KIAA1646 | |||
|
Domain Architecture |

|
|||||
| Description | Ceramide kinase (EC 2.7.1.138) (Acylsphingosine kinase) (hCERK) (Lipid kinase 4) (LK4). | |||||
|
CERK1_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 5) | NC score | 0.641539 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4Q7 | Gene names | Cerk | |||
|
Domain Architecture |
|
|||||
| Description | Ceramide kinase (EC 2.7.1.138) (Acylsphingosine kinase) (mCERK). | |||||
|
MULK_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 6) | NC score | 0.563451 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q53H12, Q75KN1, Q96GC3, Q9NP48 | Gene names | MULK, AGK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple substrate lipid kinase, mitochondrial precursor (EC 2.7.1.94) (EC 2.7.1.107) (Multi-substrate lipid kinase) (MuLK) (HsMuLK) (Acylglycerol kinase) (hAGK). | |||||
|
MULK_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 7) | NC score | 0.559769 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESW4, Q9D087 | Gene names | Mulk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple substrate lipid kinase, mitochondrial precursor (EC 2.7.1.94) (EC 2.7.1.107) (Multi-substrate lipid kinase) (MuLK). | |||||
|
CERKL_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 8) | NC score | 0.457536 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q49MI3, Q49MH9, Q49MI0, Q49MI1, Q49MI2, Q5DVJ2, Q5DVJ4, Q5DVJ5, Q6UZF6, Q6ZP59 | Gene names | CERKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ceramide kinase-like protein. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 9) | NC score | 0.081382 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.029804 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
DGKI_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.075055 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75912, Q9NZ49 | Gene names | DGKI | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase iota (EC 2.7.1.107) (Diglyceride kinase iota) (DGK-iota) (DAG kinase iota). | |||||
|
ERCC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.061408 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07992 | Gene names | ERCC1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA excision repair protein ERCC-1. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.040433 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.025609 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.051503 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
NRAM1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.033878 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41251 | Gene names | Slc11a1, Bcg, Ity, Lsh, Nramp1 | |||
|
Domain Architecture |
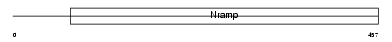
|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.056750 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.056377 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.025620 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
TNFC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.045389 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41155 | Gene names | Ltb, Tnfc, Tnfsf3 | |||
|
Domain Architecture |
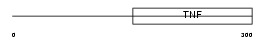
|
|||||
| Description | Lymphotoxin-beta (LT-beta) (Tumor necrosis factor C) (TNF-C) (Tumor necrosis factor ligand superfamily member 3). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.031803 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.028286 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.036923 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
DGKG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.054915 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.040014 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.003922 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.007412 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.031531 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.025961 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
RTKN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.030638 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BST9, Q8WVN1, Q96PT6, Q9HB05 | Gene names | RTKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.031428 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.050666 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
DMRTD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.042193 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGW9 | Gene names | Dmrtc2, Dmrt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2 (Doublesex- and mab-3-related transcription factor 7). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.029885 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.043093 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
WDR46_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.024806 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |
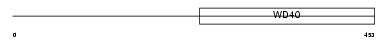
|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
CYLD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.022636 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
DGKA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.056232 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23743, O75481, O75482, O75483 | Gene names | DGKA, DAGK, DAGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.026675 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
PTN5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.009786 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
CK035_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.024279 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXW0 | Gene names | C11orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35. | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.026612 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.008317 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.018012 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.025318 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.014742 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.009856 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.022733 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.030548 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.008473 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
DEN1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.018899 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DGKZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.049649 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13574, O00542 | Gene names | DGKZ, DAGK6 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase zeta (EC 2.7.1.107) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta). | |||||
|
DGKZ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.049880 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UP3 | Gene names | Dgkz | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase zeta (EC 2.7.1.107) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta). | |||||
|
FIZ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.000194 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTJ4, Q91W14, Q9CTG3 | Gene names | Fiz1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flt3-interacting zinc finger protein 1. | |||||
|
LHX3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.008418 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |
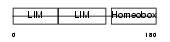
|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.023147 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BAI2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.005922 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BIM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.019410 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.009939 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.027447 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.018861 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
DGKA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.043896 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88673, Q922X2 | Gene names | Dgka, Dagk1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
IGHE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.007558 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01854 | Gene names | IGHE | |||
|
Domain Architecture |

|
|||||
| Description | Ig epsilon chain C region. | |||||
|
M3K14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.002046 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.006731 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.006133 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.010743 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.013965 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.010514 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
ARAF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.000139 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.018851 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.018117 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
DGKB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.041369 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6T7, O75241, Q75MF9, Q75MU7, Q86UI5, Q86UM9, Q9UQ29 | Gene names | DGKB, DAGK2, KIAA0718 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase beta (EC 2.7.1.107) (Diglyceride kinase beta) (DGK-beta) (DAG kinase beta) (90 kDa diacylglycerol kinase). | |||||
|
HXB4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003808 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.008853 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
KIFC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.002085 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.003141 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.010464 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.016611 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.011555 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | -0.000804 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
CXX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.050032 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15255 | Gene names | CXX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX box protein 1 (Cerebral protein 5). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.060523 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SPHK2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NRA0, Q9BRN1, Q9H0Q2, Q9NWU7 | Gene names | SPHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
SPHK2_MOUSE
|
||||||
| NC score | 0.978938 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
SPHK1_HUMAN
|
||||||
| NC score | 0.903900 (rank : 3) | θ value | 3.974e-56 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYA1, Q9HD92, Q9NY70, Q9NYL3 | Gene names | SPHK1, SPHK, SPK | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 1 (EC 2.7.1.-) (SK 1) (SPK 1). | |||||
|
CERK1_HUMAN
|
||||||
| NC score | 0.655657 (rank : 4) | θ value | 8.36355e-22 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TCT0, Q9BYB3, Q9UGE5 | Gene names | CERK, KIAA1646 | |||
|
Domain Architecture |

|
|||||
| Description | Ceramide kinase (EC 2.7.1.138) (Acylsphingosine kinase) (hCERK) (Lipid kinase 4) (LK4). | |||||
|
CERK1_MOUSE
|
||||||
| NC score | 0.641539 (rank : 5) | θ value | 1.33526e-19 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4Q7 | Gene names | Cerk | |||
|
Domain Architecture |

|
|||||
| Description | Ceramide kinase (EC 2.7.1.138) (Acylsphingosine kinase) (mCERK). | |||||
|
MULK_HUMAN
|
||||||
| NC score | 0.563451 (rank : 6) | θ value | 4.59992e-12 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q53H12, Q75KN1, Q96GC3, Q9NP48 | Gene names | MULK, AGK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple substrate lipid kinase, mitochondrial precursor (EC 2.7.1.94) (EC 2.7.1.107) (Multi-substrate lipid kinase) (MuLK) (HsMuLK) (Acylglycerol kinase) (hAGK). | |||||
|
MULK_MOUSE
|
||||||
| NC score | 0.559769 (rank : 7) | θ value | 1.33837e-11 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESW4, Q9D087 | Gene names | Mulk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple substrate lipid kinase, mitochondrial precursor (EC 2.7.1.94) (EC 2.7.1.107) (Multi-substrate lipid kinase) (MuLK). | |||||
|
CERKL_HUMAN
|
||||||
| NC score | 0.457536 (rank : 8) | θ value | 4.1701e-05 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q49MI3, Q49MH9, Q49MI0, Q49MI1, Q49MI2, Q5DVJ2, Q5DVJ4, Q5DVJ5, Q6UZF6, Q6ZP59 | Gene names | CERKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ceramide kinase-like protein. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.081382 (rank : 9) | θ value | 0.00390308 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
DGKI_HUMAN
|
||||||
| NC score | 0.075055 (rank : 10) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75912, Q9NZ49 | Gene names | DGKI | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase iota (EC 2.7.1.107) (Diglyceride kinase iota) (DGK-iota) (DAG kinase iota). | |||||
|
ERCC1_HUMAN
|
||||||
| NC score | 0.061408 (rank : 11) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07992 | Gene names | ERCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-1. | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.060523 (rank : 12) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.056750 (rank : 13) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.056377 (rank : 14) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DGKA_HUMAN
|
||||||
| NC score | 0.056232 (rank : 15) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23743, O75481, O75482, O75483 | Gene names | DGKA, DAGK, DAGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
DGKG_HUMAN
|
||||||
| NC score | 0.054915 (rank : 16) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.051503 (rank : 17) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.050666 (rank : 18) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
CXX1_HUMAN
|
||||||
| NC score | 0.050032 (rank : 19) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O15255 | Gene names | CXX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX box protein 1 (Cerebral protein 5). | |||||
|
DGKZ_MOUSE
|
||||||
| NC score | 0.049880 (rank : 20) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UP3 | Gene names | Dgkz | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase zeta (EC 2.7.1.107) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta). | |||||
|
DGKZ_HUMAN
|
||||||
| NC score | 0.049649 (rank : 21) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13574, O00542 | Gene names | DGKZ, DAGK6 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase zeta (EC 2.7.1.107) (Diglyceride kinase zeta) (DGK-zeta) (DAG kinase zeta). | |||||
|
TNFC_MOUSE
|
||||||
| NC score | 0.045389 (rank : 22) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41155 | Gene names | Ltb, Tnfc, Tnfsf3 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphotoxin-beta (LT-beta) (Tumor necrosis factor C) (TNF-C) (Tumor necrosis factor ligand superfamily member 3). | |||||
|
DGKA_MOUSE
|
||||||
| NC score | 0.043896 (rank : 23) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88673, Q922X2 | Gene names | Dgka, Dagk1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.043093 (rank : 24) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
DMRTD_MOUSE
|
||||||
| NC score | 0.042193 (rank : 25) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGW9 | Gene names | Dmrtc2, Dmrt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2 (Doublesex- and mab-3-related transcription factor 7). | |||||
|
DGKB_HUMAN
|
||||||
| NC score | 0.041369 (rank : 26) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6T7, O75241, Q75MF9, Q75MU7, Q86UI5, Q86UM9, Q9UQ29 | Gene names | DGKB, DAGK2, KIAA0718 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase beta (EC 2.7.1.107) (Diglyceride kinase beta) (DGK-beta) (DAG kinase beta) (90 kDa diacylglycerol kinase). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.040433 (rank : 27) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.040014 (rank : 28) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.036923 (rank : 29) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
NRAM1_MOUSE
|
||||||
| NC score | 0.033878 (rank : 30) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41251 | Gene names | Slc11a1, Bcg, Ity, Lsh, Nramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.031803 (rank : 31) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.031531 (rank : 32) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.031428 (rank : 33) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
RTKN_HUMAN
|
||||||
| NC score | 0.030638 (rank : 34) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BST9, Q8WVN1, Q96PT6, Q9HB05 | Gene names | RTKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.030548 (rank : 35) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.029885 (rank : 36) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.029804 (rank : 37) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.028286 (rank : 38) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.027447 (rank : 39) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.026675 (rank : 40) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.026612 (rank : 41) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.025961 (rank : 42) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.025620 (rank : 43) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.025609 (rank : 44) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.025318 (rank : 45) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WDR46_MOUSE
|
||||||
| NC score | 0.024806 (rank : 46) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
CK035_HUMAN
|
||||||
| NC score | 0.024279 (rank : 47) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXW0 | Gene names | C11orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.023147 (rank : 48) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.022733 (rank : 49) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CYLD_MOUSE
|
||||||
| NC score | 0.022636 (rank : 50) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
BIM_HUMAN
|
||||||
| NC score | 0.019410 (rank : 51) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
DEN1A_MOUSE
|
||||||
| NC score | 0.018899 (rank : 52) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.018861 (rank : 53) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.018851 (rank : 54) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.018117 (rank : 55) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.018012 (rank : 56) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.016611 (rank : 57) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.014742 (rank : 58) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.013965 (rank : 59) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.011555 (rank : 60) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.010743 (rank : 61) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.010514 (rank : 62) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.010464 (rank : 63) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.009939 (rank : 64) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.009856 (rank : 65) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.009786 (rank : 66) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.008853 (rank : 67) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.008473 (rank : 68) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
LHX3_HUMAN
|
||||||
| NC score | 0.008418 (rank : 69) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.008317 (rank : 70) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
IGHE_HUMAN
|
||||||
| NC score | 0.007558 (rank : 71) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01854 | Gene names | IGHE | |||
|
Domain Architecture |

|
|||||
| Description | Ig epsilon chain C region. | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.007412 (rank : 72) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.006731 (rank : 73) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.006133 (rank : 74) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
BAI2_HUMAN
|
||||||
| NC score | 0.005922 (rank : 75) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.003922 (rank : 76) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
HXB4_MOUSE
|
||||||
| NC score | 0.003808 (rank : 77) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.003141 (rank : 78) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
KIFC3_HUMAN
|
||||||
| NC score | 0.002085 (rank : 79) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
M3K14_MOUSE
|
||||||
| NC score | 0.002046 (rank : 80) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
FIZ1_MOUSE
|
||||||
| NC score | 0.000194 (rank : 81) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTJ4, Q91W14, Q9CTG3 | Gene names | Fiz1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flt3-interacting zinc finger protein 1. | |||||
|
ARAF_HUMAN
|
||||||
| NC score | 0.000139 (rank : 82) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000804 (rank : 83) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||