Please be patient as the page loads
|
MPRI_HUMAN
|
||||||
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MPRI_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
MPRI_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992617 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
MMP2_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 3) | NC score | 0.262698 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 4) | NC score | 0.259677 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 5) | NC score | 0.284082 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.284525 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 7) | NC score | 0.436494 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 8) | NC score | 0.433332 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 9) | NC score | 0.120019 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
FINC_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 10) | NC score | 0.220719 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 11) | NC score | 0.220257 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
HGFA_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 12) | NC score | 0.118973 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
MRC1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 13) | NC score | 0.261892 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MPRD_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 14) | NC score | 0.255759 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20645 | Gene names | M6PR, MPR46, MPRD | |||
|
Domain Architecture |
|
|||||
| Description | Cation-dependent mannose-6-phosphate receptor precursor (CD Man-6-P receptor) (CD-MPR) (46 kDa mannose 6-phosphate receptor) (MPR 46). | |||||
|
MPRD_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 15) | NC score | 0.266868 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24668 | Gene names | M6pr, 46mpr | |||
|
Domain Architecture |
|
|||||
| Description | Cation-dependent mannose-6-phosphate receptor precursor (CD Man-6-P receptor) (CD-MPR) (46 kDa mannose 6-phosphate receptor) (MPR 46). | |||||
|
LY75_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 16) | NC score | 0.262225 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 17) | NC score | 0.253003 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
FA12_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.114623 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
MRC2_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.251619 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 20) | NC score | 0.252562 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
LY75_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 21) | NC score | 0.252683 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.028594 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SL9A5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.033285 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.033957 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
MYBB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.023236 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
ELOA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.024177 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.020037 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
3HAO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.032771 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P46952, Q53QZ7, Q8N6N9 | Gene names | HAAO | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyanthranilate 3,4-dioxygenase (EC 1.13.11.6) (3-HAO) (3- hydroxyanthranilic acid dioxygenase) (3-hydroxyanthranilate oxygenase). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.013983 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.013550 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.013776 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.019970 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.046610 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.015299 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
EAN57_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.024216 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.012042 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
PTEN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.021299 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
PTEN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.021298 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.024900 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SGCD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.015137 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P82347 | Gene names | Sgcd | |||
|
Domain Architecture |

|
|||||
| Description | Delta-sarcoglycan (SG-delta) (35 kDa dystrophin-associated glycoprotein) (35DAG). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.008967 (rank : 74) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
GNPTG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.022927 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6S5C2, Q7TNE0, Q8C5J3 | Gene names | Gnptg, Mdcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunit gamma precursor (GlcNAc-1-phosphotransferase subunit gamma) (UDP-N-acetylglucosamine- 1-phosphotransferase, subunit gamma) (M6PR domain-containing protein 1). | |||||
|
GRP78_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.014886 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.014844 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
LRBA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.020785 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.025678 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
DMXL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.008071 (rank : 75) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.042850 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.012186 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.009747 (rank : 72) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
FETUB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.011993 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |
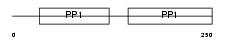
|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.006584 (rank : 77) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.009086 (rank : 73) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.009829 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.011981 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.012254 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.012268 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYOM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.019104 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52179 | Gene names | MYOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (190 kDa titin-associated protein) (190 kDa connectin- associated protein). | |||||
|
OR2Z1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | -0.001679 (rank : 85) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG97, Q6IFK0, Q96R25 | Gene names | OR2Z1, OR2Z2 | |||
|
Domain Architecture |
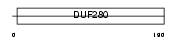
|
|||||
| Description | Olfactory receptor 2Z1 (Olfactory receptor OR19-4). | |||||
|
PHLPL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.003103 (rank : 84) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PTN6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.003339 (rank : 83) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.004122 (rank : 82) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
RARG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.006145 (rank : 79) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.006160 (rank : 78) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.006109 (rank : 81) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.006119 (rank : 80) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.007234 (rank : 76) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
CD302_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.073221 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.072312 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CHODL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056023 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.056055 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
EMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050664 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
MMP10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050532 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051099 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.051524 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.051746 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051904 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051285 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.050493 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.053759 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.050257 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.050246 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.058284 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.058292 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
PRG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.051456 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
MPRI_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
MPRI_MOUSE
|
||||||
| NC score | 0.992617 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.436494 (rank : 3) | θ value | 3.64472e-09 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.433332 (rank : 4) | θ value | 3.64472e-09 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.284525 (rank : 5) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.284082 (rank : 6) | θ value | 2.13673e-09 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MPRD_MOUSE
|
||||||
| NC score | 0.266868 (rank : 7) | θ value | 2.88788e-06 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24668 | Gene names | M6pr, 46mpr | |||
|
Domain Architecture |
|
|||||
| Description | Cation-dependent mannose-6-phosphate receptor precursor (CD Man-6-P receptor) (CD-MPR) (46 kDa mannose 6-phosphate receptor) (MPR 46). | |||||
|
MMP2_HUMAN
|
||||||
| NC score | 0.262698 (rank : 8) | θ value | 1.47974e-10 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
LY75_HUMAN
|
||||||
| NC score | 0.262225 (rank : 9) | θ value | 3.77169e-06 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| NC score | 0.261892 (rank : 10) | θ value | 2.21117e-06 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MMP2_MOUSE
|
||||||
| NC score | 0.259677 (rank : 11) | θ value | 7.34386e-10 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MPRD_HUMAN
|
||||||
| NC score | 0.255759 (rank : 12) | θ value | 2.88788e-06 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20645 | Gene names | M6PR, MPR46, MPRD | |||
|
Domain Architecture |
|
|||||
| Description | Cation-dependent mannose-6-phosphate receptor precursor (CD Man-6-P receptor) (CD-MPR) (46 kDa mannose 6-phosphate receptor) (MPR 46). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.253003 (rank : 13) | θ value | 8.40245e-06 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.252683 (rank : 14) | θ value | 5.44631e-05 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.252562 (rank : 15) | θ value | 1.43324e-05 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.251619 (rank : 16) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.220719 (rank : 17) | θ value | 1.17247e-07 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.220257 (rank : 18) | θ value | 1.17247e-07 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.120019 (rank : 19) | θ value | 4.0297e-08 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.118973 (rank : 20) | θ value | 1.17247e-07 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.114623 (rank : 21) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
CD302_HUMAN
|
||||||
| NC score | 0.073221 (rank : 22) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.072312 (rank : 23) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
MMP7_MOUSE
|
||||||
| NC score | 0.058292 (rank : 24) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_HUMAN
|
||||||
| NC score | 0.058284 (rank : 25) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
CHODL_MOUSE
|
||||||
| NC score | 0.056055 (rank : 26) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_HUMAN
|
||||||
| NC score | 0.056023 (rank : 27) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
MMP26_HUMAN
|
||||||
| NC score | 0.053759 (rank : 28) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.051904 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.051746 (rank : 30) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.051524 (rank : 31) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.051456 (rank : 32) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
MMP13_MOUSE
|
||||||
| NC score | 0.051285 (rank : 33) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.051099 (rank : 34) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
EMBP_MOUSE
|
||||||
| NC score | 0.050664 (rank : 35) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
MMP10_HUMAN
|
||||||
| NC score | 0.050532 (rank : 36) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP1_HUMAN
|
||||||
| NC score | 0.050493 (rank : 37) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP3_HUMAN
|
||||||
| NC score | 0.050257 (rank : 38) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP3_MOUSE
|
||||||
| NC score | 0.050246 (rank : 39) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.046610 (rank : 40) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.042850 (rank : 41) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.033957 (rank : 42) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
SL9A5_HUMAN
|
||||||
| NC score | 0.033285 (rank : 43) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
3HAO_HUMAN
|
||||||
| NC score | 0.032771 (rank : 44) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P46952, Q53QZ7, Q8N6N9 | Gene names | HAAO | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyanthranilate 3,4-dioxygenase (EC 1.13.11.6) (3-HAO) (3- hydroxyanthranilic acid dioxygenase) (3-hydroxyanthranilate oxygenase). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.028594 (rank : 45) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.025678 (rank : 46) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.024900 (rank : 47) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
EAN57_HUMAN
|
||||||
| NC score | 0.024216 (rank : 48) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
ELOA1_MOUSE
|
||||||
| NC score | 0.024177 (rank : 49) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MYBB_HUMAN
|
||||||
| NC score | 0.023236 (rank : 50) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
GNPTG_MOUSE
|
||||||
| NC score | 0.022927 (rank : 51) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6S5C2, Q7TNE0, Q8C5J3 | Gene names | Gnptg, Mdcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunit gamma precursor (GlcNAc-1-phosphotransferase subunit gamma) (UDP-N-acetylglucosamine- 1-phosphotransferase, subunit gamma) (M6PR domain-containing protein 1). | |||||
|
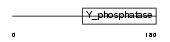
PTEN_HUMAN
|
||||||
| NC score | 0.021299 (rank : 52) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
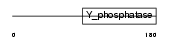
PTEN_MOUSE
|
||||||
| NC score | 0.021298 (rank : 53) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.020785 (rank : 54) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.020037 (rank : 55) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.019970 (rank : 56) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MYOM1_HUMAN
|
||||||
| NC score | 0.019104 (rank : 57) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52179 | Gene names | MYOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (190 kDa titin-associated protein) (190 kDa connectin- associated protein). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.015299 (rank : 58) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
SGCD_MOUSE
|
||||||
| NC score | 0.015137 (rank : 59) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P82347 | Gene names | Sgcd | |||
|
Domain Architecture |

|
|||||
| Description | Delta-sarcoglycan (SG-delta) (35 kDa dystrophin-associated glycoprotein) (35DAG). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.014886 (rank : 60) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.014844 (rank : 61) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.013983 (rank : 62) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.013776 (rank : 63) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.013550 (rank : 64) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.012268 (rank : 65) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.012254 (rank : 66) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.012186 (rank : 67) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.012042 (rank : 68) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
FETUB_HUMAN
|
||||||
| NC score | 0.011993 (rank : 69) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.011981 (rank : 70) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.009829 (rank : 71) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.009747 (rank : 72) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.009086 (rank : 73) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.008967 (rank : 74) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
DMXL2_HUMAN
|
||||||
| NC score | 0.008071 (rank : 75) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.007234 (rank : 76) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.006584 (rank : 77) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RARG1_MOUSE
|
||||||
| NC score | 0.006160 (rank : 78) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG1_HUMAN
|
||||||
| NC score | 0.006145 (rank : 79) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG2_MOUSE
|
||||||
| NC score | 0.006119 (rank : 80) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RARG2_HUMAN
|
||||||
| NC score | 0.006109 (rank : 81) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.004122 (rank : 82) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTN6_MOUSE
|
||||||
| NC score | 0.003339 (rank : 83) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PHLPL_MOUSE
|
||||||
| NC score | 0.003103 (rank : 84) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
OR2Z1_HUMAN
|
||||||
| NC score | -0.001679 (rank : 85) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG97, Q6IFK0, Q96R25 | Gene names | OR2Z1, OR2Z2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 2Z1 (Olfactory receptor OR19-4). | |||||