Please be patient as the page loads
|
FETUB_HUMAN
|
||||||
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |
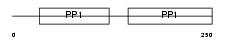
|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FETUB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
FETUB_MOUSE
|
||||||
| θ value | 4.15819e-130 (rank : 2) | NC score | 0.964460 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXC1 | Gene names | Fetub | |||
|
Domain Architecture |
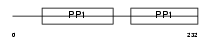
|
|||||
| Description | Fetuin-B precursor (IRL685). | |||||
|
FETUA_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 3) | NC score | 0.654558 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02765, O14961, O14962, Q9P152 | Gene names | AHSG, FETUA | |||
|
Domain Architecture |
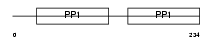
|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Alpha-2-Z-globulin) (Ba- alpha-2-glycoprotein) [Contains: Alpha-2-HS-glycoprotein chain A; Alpha-2-HS-glycoprotein chain B]. | |||||
|
FETUA_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 4) | NC score | 0.650557 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29699, O35634 | Gene names | Ahsg, Fetua | |||
|
Domain Architecture |
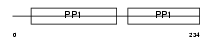
|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Countertrypin). | |||||
|
HRG_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 5) | NC score | 0.523275 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |
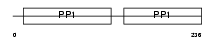
|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
KNG1_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 6) | NC score | 0.483775 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08677, O08676, Q91XK5 | Gene names | Kng1, Kng | |||
|
Domain Architecture |
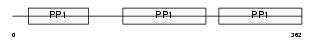
|
|||||
| Description | Kininogen-1 precursor [Contains: Kininogen-1 heavy chain; Bradykinin; Kininogen-1 light chain]. | |||||
|
KNG1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 7) | NC score | 0.474625 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P01042, P01043 | Gene names | KNG1, BDK, KNG | |||
|
Domain Architecture |
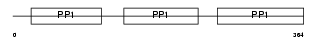
|
|||||
| Description | Kininogen-1 precursor (Alpha-2-thiol proteinase inhibitor) [Contains: Kininogen-1 heavy chain; Bradykinin (Kallidin I); Lysyl-bradykinin (Kallidin II); Kininogen-1 light chain; Low molecular weight growth- promoting factor]. | |||||
|
HLF_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.081549 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
CYTF_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.205182 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76096, Q7Z4J8, Q9UED4 | Gene names | CST7 | |||
|
Domain Architecture |
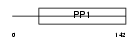
|
|||||
| Description | Cystatin F precursor (Leukocystatin) (Cystatin-7) (Cystatin-like metastasis-associated protein) (CMAP). | |||||
|
CYTC_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.144338 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21460, Q544Y0 | Gene names | Cst3 | |||
|
Domain Architecture |
|
|||||
| Description | Cystatin C precursor (Cystatin 3). | |||||
|
HLF_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.076818 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
SPP24_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.083496 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1I3, Q9DCG1 | Gene names | Spp2, Spp24 | |||
|
Domain Architecture |
|
|||||
| Description | Secreted phosphoprotein 24 precursor (Spp-24) (Secreted phosphoprotein 2). | |||||
|
CYTN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.114599 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P01037, Q96LE6, Q9UCQ6 | Gene names | CST1 | |||
|
Domain Architecture |
|
|||||
| Description | Cystatin SN precursor (Salivary cystatin SA-1) (Cystain SA-I). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.013896 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.014029 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
CYTC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.121401 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P01034 | Gene names | CST3 | |||
|
Domain Architecture |
|
|||||
| Description | Cystatin C precursor (Neuroendocrine basic polypeptide) (Gamma-trace) (Post-gamma-globulin). | |||||
|
CYTS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.101200 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01036, Q9UBI5, Q9UCS9 | Gene names | CST4 | |||
|
Domain Architecture |
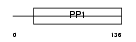
|
|||||
| Description | Cystatin S precursor (Salivary acidic protein 1) (Cystatin SA-III). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.017077 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
TSYL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.022303 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |
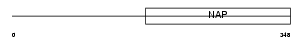
|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.010927 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.013942 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.009809 (rank : 38) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
CYTD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.093685 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28325, Q5JRF5 | Gene names | CST5 | |||
|
Domain Architecture |
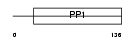
|
|||||
| Description | Cystatin D precursor. | |||||
|
CYTT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.116017 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P09228, Q9UCQ7 | Gene names | CST2 | |||
|
Domain Architecture |
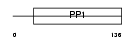
|
|||||
| Description | Cystatin SA precursor (Cystatin S5). | |||||
|
5NT1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.017171 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YE9, Q91Y48 | Gene names | Nt5c1b, Airp | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.017352 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.010043 (rank : 37) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CA106_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.013833 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
FANCG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.016171 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
|
MPRI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.011993 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
NMI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.014864 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13287, Q53TI8, Q9BVE5 | Gene names | NMI | |||
|
Domain Architecture |
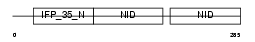
|
|||||
| Description | N-myc-interactor (Nmi) (N-myc and STAT interactor). | |||||
|
CST11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.061499 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H112, Q8WXU5, Q8WXU6, Q9H113 | Gene names | CST11, CST8L | |||
|
Domain Architecture |
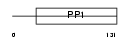
|
|||||
| Description | Cystatin-11 precursor. | |||||
|
CST11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.064249 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D269 | Gene names | Cst11 | |||
|
Domain Architecture |
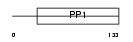
|
|||||
| Description | Cystatin-11 precursor. | |||||
|
CST13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.059050 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80ZN5, Q9DAP1 | Gene names | Cst13, Cymg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cystatin-13 precursor (Cystatin T). | |||||
|
CST1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.089187 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H114, Q64FF7 | Gene names | CSTL1 | |||
|
Domain Architecture |
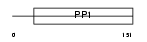
|
|||||
| Description | Cystatin-like 1 precursor (RCET11). | |||||
|
CST8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.062639 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60676, Q2M2X6 | Gene names | CST8, CRES | |||
|
Domain Architecture |
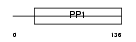
|
|||||
| Description | Cystatin-related epididymal spermatogenic protein precursor (Cystatin- 8). | |||||
|
CYTF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.164693 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O89098 | Gene names | Cst7 | |||
|
Domain Architecture |
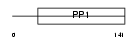
|
|||||
| Description | Cystatin F precursor (Leukocystatin) (Cystatin-7) (Cystatin-like metastasis-associated protein) (CMAP). | |||||
|
CYTM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.108150 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15828 | Gene names | CST6 | |||
|
Domain Architecture |
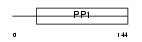
|
|||||
| Description | Cystatin M precursor (Cystatin E). | |||||
|
FETUB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
FETUB_MOUSE
|
||||||
| NC score | 0.964460 (rank : 2) | θ value | 4.15819e-130 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXC1 | Gene names | Fetub | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (IRL685). | |||||
|
FETUA_HUMAN
|
||||||
| NC score | 0.654558 (rank : 3) | θ value | 9.56915e-18 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02765, O14961, O14962, Q9P152 | Gene names | AHSG, FETUA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Alpha-2-Z-globulin) (Ba- alpha-2-glycoprotein) [Contains: Alpha-2-HS-glycoprotein chain A; Alpha-2-HS-glycoprotein chain B]. | |||||
|
FETUA_MOUSE
|
||||||
| NC score | 0.650557 (rank : 4) | θ value | 1.24977e-17 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29699, O35634 | Gene names | Ahsg, Fetua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Countertrypin). | |||||
|
HRG_HUMAN
|
||||||
| NC score | 0.523275 (rank : 5) | θ value | 9.90251e-15 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |

|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
KNG1_MOUSE
|
||||||
| NC score | 0.483775 (rank : 6) | θ value | 1.74796e-11 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08677, O08676, Q91XK5 | Gene names | Kng1, Kng | |||
|
Domain Architecture |

|
|||||
| Description | Kininogen-1 precursor [Contains: Kininogen-1 heavy chain; Bradykinin; Kininogen-1 light chain]. | |||||
|
KNG1_HUMAN
|
||||||
| NC score | 0.474625 (rank : 7) | θ value | 3.29651e-10 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P01042, P01043 | Gene names | KNG1, BDK, KNG | |||
|
Domain Architecture |

|
|||||
| Description | Kininogen-1 precursor (Alpha-2-thiol proteinase inhibitor) [Contains: Kininogen-1 heavy chain; Bradykinin (Kallidin I); Lysyl-bradykinin (Kallidin II); Kininogen-1 light chain; Low molecular weight growth- promoting factor]. | |||||
|
CYTF_HUMAN
|
||||||
| NC score | 0.205182 (rank : 8) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76096, Q7Z4J8, Q9UED4 | Gene names | CST7 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin F precursor (Leukocystatin) (Cystatin-7) (Cystatin-like metastasis-associated protein) (CMAP). | |||||
|
CYTF_MOUSE
|
||||||
| NC score | 0.164693 (rank : 9) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O89098 | Gene names | Cst7 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin F precursor (Leukocystatin) (Cystatin-7) (Cystatin-like metastasis-associated protein) (CMAP). | |||||
|
CYTC_MOUSE
|
||||||
| NC score | 0.144338 (rank : 10) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21460, Q544Y0 | Gene names | Cst3 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin C precursor (Cystatin 3). | |||||
|
CYTC_HUMAN
|
||||||
| NC score | 0.121401 (rank : 11) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P01034 | Gene names | CST3 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin C precursor (Neuroendocrine basic polypeptide) (Gamma-trace) (Post-gamma-globulin). | |||||
|
CYTT_HUMAN
|
||||||
| NC score | 0.116017 (rank : 12) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P09228, Q9UCQ7 | Gene names | CST2 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin SA precursor (Cystatin S5). | |||||
|
CYTN_HUMAN
|
||||||
| NC score | 0.114599 (rank : 13) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P01037, Q96LE6, Q9UCQ6 | Gene names | CST1 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin SN precursor (Salivary cystatin SA-1) (Cystain SA-I). | |||||
|
CYTM_HUMAN
|
||||||
| NC score | 0.108150 (rank : 14) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15828 | Gene names | CST6 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin M precursor (Cystatin E). | |||||
|
CYTS_HUMAN
|
||||||
| NC score | 0.101200 (rank : 15) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01036, Q9UBI5, Q9UCS9 | Gene names | CST4 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin S precursor (Salivary acidic protein 1) (Cystatin SA-III). | |||||
|
CYTD_HUMAN
|
||||||
| NC score | 0.093685 (rank : 16) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28325, Q5JRF5 | Gene names | CST5 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin D precursor. | |||||
|
CST1L_HUMAN
|
||||||
| NC score | 0.089187 (rank : 17) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H114, Q64FF7 | Gene names | CSTL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin-like 1 precursor (RCET11). | |||||
|
SPP24_MOUSE
|
||||||
| NC score | 0.083496 (rank : 18) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1I3, Q9DCG1 | Gene names | Spp2, Spp24 | |||
|
Domain Architecture |

|
|||||
| Description | Secreted phosphoprotein 24 precursor (Spp-24) (Secreted phosphoprotein 2). | |||||
|
HLF_MOUSE
|
||||||
| NC score | 0.081549 (rank : 19) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_HUMAN
|
||||||
| NC score | 0.076818 (rank : 20) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
CST11_MOUSE
|
||||||
| NC score | 0.064249 (rank : 21) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D269 | Gene names | Cst11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin-11 precursor. | |||||
|
CST8_HUMAN
|
||||||
| NC score | 0.062639 (rank : 22) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60676, Q2M2X6 | Gene names | CST8, CRES | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin-related epididymal spermatogenic protein precursor (Cystatin- 8). | |||||
|
CST11_HUMAN
|
||||||
| NC score | 0.061499 (rank : 23) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H112, Q8WXU5, Q8WXU6, Q9H113 | Gene names | CST11, CST8L | |||
|
Domain Architecture |

|
|||||
| Description | Cystatin-11 precursor. | |||||
|
CST13_MOUSE
|
||||||
| NC score | 0.059050 (rank : 24) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80ZN5, Q9DAP1 | Gene names | Cst13, Cymg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cystatin-13 precursor (Cystatin T). | |||||
|
TSYL1_MOUSE
|
||||||
| NC score | 0.022303 (rank : 25) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.017352 (rank : 26) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
5NT1B_MOUSE
|
||||||
| NC score | 0.017171 (rank : 27) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YE9, Q91Y48 | Gene names | Nt5c1b, Airp | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.017077 (rank : 28) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
FANCG_HUMAN
|
||||||
| NC score | 0.016171 (rank : 29) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
|
NMI_HUMAN
|
||||||
| NC score | 0.014864 (rank : 30) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13287, Q53TI8, Q9BVE5 | Gene names | NMI | |||
|
Domain Architecture |

|
|||||
| Description | N-myc-interactor (Nmi) (N-myc and STAT interactor). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.014029 (rank : 31) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.013942 (rank : 32) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.013896 (rank : 33) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.013833 (rank : 34) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
MPRI_HUMAN
|
||||||
| NC score | 0.011993 (rank : 35) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.010927 (rank : 36) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.010043 (rank : 37) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.009809 (rank : 38) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||