Please be patient as the page loads
|
FANCG_HUMAN
|
||||||
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FANCG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
|
FANCG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982915 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQR6 | Gene names | Fancg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein homolog (Protein FACG). | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 3) | NC score | 0.103011 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 4) | NC score | 0.148227 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.094587 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.082341 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.079991 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ANXA4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.018112 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09525, Q96F33, Q9BWK1 | Gene names | ANXA4, ANX4 | |||
|
Domain Architecture |
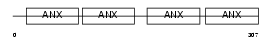
|
|||||
| Description | Annexin A4 (Annexin IV) (Lipocortin IV) (Endonexin I) (Chromobindin-4) (Protein II) (P32.5) (Placental anticoagulant protein II) (PAP-II) (PP4-X) (35-beta calcimedin) (Carbohydrate-binding protein P33/P41) (P33/41). | |||||
|
ERCC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.041079 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ERCC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.041005 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ANXA4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.016623 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97429 | Gene names | Anxa4, Anx4 | |||
|
Domain Architecture |
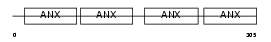
|
|||||
| Description | Annexin A4 (Annexin IV). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.019830 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.017456 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.007293 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.006559 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
PIAS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.010989 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
FETUB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.016171 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |
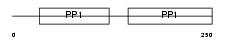
|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
FANCG_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15287 | Gene names | FANCG, XRCC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein (Protein FACG) (DNA-repair protein XRCC9). | |||||
|
FANCG_MOUSE
|
||||||
| NC score | 0.982915 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQR6 | Gene names | Fancg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein homolog (Protein FACG). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.148227 (rank : 3) | θ value | 0.0193708 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.103011 (rank : 4) | θ value | 0.00869519 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.094587 (rank : 5) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.082341 (rank : 6) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.079991 (rank : 7) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ERCC2_HUMAN
|
||||||
| NC score | 0.041079 (rank : 8) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ERCC2_MOUSE
|
||||||
| NC score | 0.041005 (rank : 9) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.019830 (rank : 10) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ANXA4_HUMAN
|
||||||
| NC score | 0.018112 (rank : 11) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09525, Q96F33, Q9BWK1 | Gene names | ANXA4, ANX4 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A4 (Annexin IV) (Lipocortin IV) (Endonexin I) (Chromobindin-4) (Protein II) (P32.5) (Placental anticoagulant protein II) (PAP-II) (PP4-X) (35-beta calcimedin) (Carbohydrate-binding protein P33/P41) (P33/41). | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.017456 (rank : 12) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
ANXA4_MOUSE
|
||||||
| NC score | 0.016623 (rank : 13) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97429 | Gene names | Anxa4, Anx4 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A4 (Annexin IV). | |||||
|
FETUB_HUMAN
|
||||||
| NC score | 0.016171 (rank : 14) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGM5, Q1RMZ0, Q6DK58, Q6GRB6, Q9Y6Z0 | Gene names | FETUB | |||
|
Domain Architecture |

|
|||||
| Description | Fetuin-B precursor (Gugu) (IRL685) (16G2). | |||||
|
PIAS2_HUMAN
|
||||||
| NC score | 0.010989 (rank : 15) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.007293 (rank : 16) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.006559 (rank : 17) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||