Please be patient as the page loads
|
ODP2_HUMAN
|
||||||
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ODP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODPX_HUMAN
|
||||||
| θ value | 1.8332e-77 (rank : 2) | NC score | 0.927740 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 1.31376e-75 (rank : 3) | NC score | 0.925731 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODB2_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 4) | NC score | 0.819059 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
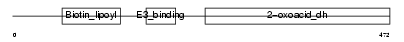
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 5) | NC score | 0.810752 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
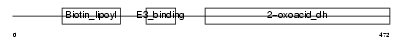
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 6) | NC score | 0.765455 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 7) | NC score | 0.759598 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.037320 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.044054 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ZF276_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.009611 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.030857 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.026460 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
GT2D1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.039773 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
PDLI7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.022221 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.022282 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.025577 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.041122 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.034897 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.016545 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.044289 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
UROL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.024975 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
PSMF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.035671 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92530, Q9H4I1 | Gene names | PSMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit (hPI31). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.030542 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.037380 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CD45_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.009374 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.019096 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.023905 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.026125 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.032229 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.026255 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.037826 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.023223 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HBS1L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.015158 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.024757 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PNMA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.018119 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHK0 | Gene names | Pnma2 | |||
|
Domain Architecture |

|
|||||
| Description | Paraneoplastic antigen Ma2 homolog. | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.029789 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.017618 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.013816 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.004669 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.020765 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.023563 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.004719 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
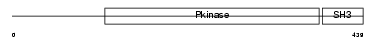
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.027549 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.030526 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.009433 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.014837 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.011643 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.006257 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.025606 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.013355 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.018624 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CS021_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.018313 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IVT2 | Gene names | C19orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.020788 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.018307 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.020796 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.020778 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ICB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.011659 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TEJ8, O60560, Q5TEJ1, Q5TEJ9, Q5TEK1, Q68DP4, Q9BYB6, Q9NS90 | Gene names | ICB1, C1orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein (Protein ICB-1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.020559 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.007963 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODPX_HUMAN
|
||||||
| NC score | 0.927740 (rank : 2) | θ value | 1.8332e-77 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.925731 (rank : 3) | θ value | 1.31376e-75 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.819059 (rank : 4) | θ value | 2.26708e-35 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.810752 (rank : 5) | θ value | 4.42448e-31 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.765455 (rank : 6) | θ value | 1.37858e-24 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.759598 (rank : 7) | θ value | 3.07116e-24 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.044289 (rank : 8) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.044054 (rank : 9) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.041122 (rank : 10) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
GT2D1_MOUSE
|
||||||
| NC score | 0.039773 (rank : 11) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.037826 (rank : 12) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.037380 (rank : 13) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.037320 (rank : 14) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
PSMF1_HUMAN
|
||||||
| NC score | 0.035671 (rank : 15) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92530, Q9H4I1 | Gene names | PSMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit (hPI31). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.034897 (rank : 16) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.032229 (rank : 17) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.030857 (rank : 18) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.030542 (rank : 19) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.030526 (rank : 20) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.029789 (rank : 21) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.027549 (rank : 22) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.026460 (rank : 23) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.026255 (rank : 24) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.026125 (rank : 25) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.025606 (rank : 26) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.025577 (rank : 27) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
UROL1_MOUSE
|
||||||
| NC score | 0.024975 (rank : 28) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.024757 (rank : 29) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.023905 (rank : 30) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.023563 (rank : 31) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.023223 (rank : 32) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.022282 (rank : 33) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PDLI7_MOUSE
|
||||||
| NC score | 0.022221 (rank : 34) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.020796 (rank : 35) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.020788 (rank : 36) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.020778 (rank : 37) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.020765 (rank : 38) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.020559 (rank : 39) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.019096 (rank : 40) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.018624 (rank : 41) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CS021_HUMAN
|
||||||
| NC score | 0.018313 (rank : 42) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IVT2 | Gene names | C19orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.018307 (rank : 43) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PNMA2_MOUSE
|
||||||
| NC score | 0.018119 (rank : 44) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHK0 | Gene names | Pnma2 | |||
|
Domain Architecture |

|
|||||
| Description | Paraneoplastic antigen Ma2 homolog. | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.017618 (rank : 45) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.016545 (rank : 46) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
HBS1L_MOUSE
|
||||||
| NC score | 0.015158 (rank : 47) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q69ZS7, Q91Z32, Q9CVT2, Q9CZ95, Q9WTY5 | Gene names | Hbs1l, Hbs1, Kiaa1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.014837 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.013816 (rank : 49) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.013355 (rank : 50) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ICB1_HUMAN
|
||||||
| NC score | 0.011659 (rank : 51) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TEJ8, O60560, Q5TEJ1, Q5TEJ9, Q5TEK1, Q68DP4, Q9BYB6, Q9NS90 | Gene names | ICB1, C1orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein (Protein ICB-1). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.011643 (rank : 52) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
ZF276_HUMAN
|
||||||
| NC score | 0.009611 (rank : 53) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.009433 (rank : 54) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.009374 (rank : 55) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.007963 (rank : 56) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.006257 (rank : 57) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.004719 (rank : 58) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.004669 (rank : 59) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||