Please be patient as the page loads
|
MOT8_MOUSE
|
||||||
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MOT8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984316 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MOT8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MOT2_HUMAN
|
||||||
| θ value | 3.16345e-37 (rank : 3) | NC score | 0.849903 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60669, Q9UPB3 | Gene names | SLC16A7, MCT2 | |||
|
Domain Architecture |
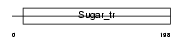
|
|||||
| Description | Monocarboxylate transporter 2 (MCT 2) (Solute carrier family 16 member 7). | |||||
|
MOT4_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 4) | NC score | 0.857556 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P57787, Q9ES80, Q9ESF8 | Gene names | Slc16a3, Mct4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 4 (MCT 4) (Solute carrier family 16 member 3). | |||||
|
MOT4_HUMAN
|
||||||
| θ value | 7.04741e-37 (rank : 5) | NC score | 0.857450 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15427 | Gene names | SLC16A3, MCT3, MCT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 4 (MCT 4) (MCT 3) (Solute carrier family 16 member 3). | |||||
|
MOT3_MOUSE
|
||||||
| θ value | 9.20422e-37 (rank : 6) | NC score | 0.856773 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35308 | Gene names | Slc16a8, Mct3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 3 (MCT 3) (Solute carrier family 16 member 8) (Proton-coupled monocarboxylate transporter 3). | |||||
|
MOT3_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 7) | NC score | 0.852520 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95907, Q9UBE2 | Gene names | SLC16A8, MCT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 3 (MCT 3) (Solute carrier family 16 member 8). | |||||
|
MOT1_HUMAN
|
||||||
| θ value | 2.34606e-32 (rank : 8) | NC score | 0.841565 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53985, Q9NSJ9 | Gene names | SLC16A1, MCT1 | |||
|
Domain Architecture |
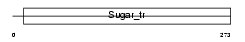
|
|||||
| Description | Monocarboxylate transporter 1 (MCT 1) (Solute carrier family 16 member 1). | |||||
|
MOT2_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 9) | NC score | 0.843236 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70451 | Gene names | Slc16a7, Mct2 | |||
|
Domain Architecture |
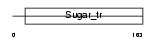
|
|||||
| Description | Monocarboxylate transporter 2 (MCT 2) (Solute carrier family 16 member 7). | |||||
|
MOT1_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 10) | NC score | 0.838783 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P53986 | Gene names | Slc16a1, Mct1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 1 (MCT 1) (Solute carrier family 16 member 1). | |||||
|
MOT6_HUMAN
|
||||||
| θ value | 9.2256e-29 (rank : 11) | NC score | 0.846490 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15375 | Gene names | SLC16A5, MCT5, MCT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 6 (MCT 6) (MCT 5) (Solute carrier family 16 member 5). | |||||
|
MOT7_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 12) | NC score | 0.799090 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15403 | Gene names | SLC16A6, MCT6, MCT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 7 (MCT 7) (MCT 6) (Solute carrier family 16 member 6). | |||||
|
MOT5_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 13) | NC score | 0.779393 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15374 | Gene names | SLC16A4, MCT4, MCT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 5 (MCT 5) (MCT 4) (Solute carrier family 16 member 4). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 14) | NC score | 0.071867 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 15) | NC score | 0.059813 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 16) | NC score | 0.086100 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 17) | NC score | 0.055089 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.108352 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 19) | NC score | 0.058825 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 20) | NC score | 0.081523 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
FLVC2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 21) | NC score | 0.095020 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91X85 | Gene names | Flvcr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.056813 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.073319 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.048220 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.036077 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.077053 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.048303 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SRC8_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.025838 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
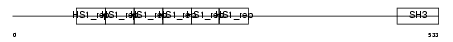
|
|||||
| Description | Src substrate cortactin. | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.082536 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.065195 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.036704 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.026277 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.037902 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.024546 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.039953 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.045803 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
S17A5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.037532 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRA2, Q8NBR5, Q9UGH0 | Gene names | SLC17A5 | |||
|
Domain Architecture |
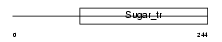
|
|||||
| Description | Sialin (Solute carrier family 17 member 5) (Sodium/sialic acid cotransporter) (AST) (Membrane glycoprotein HP59). | |||||
|
VACHT_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.075100 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35304 | Gene names | Slc18a3, Vacht | |||
|
Domain Architecture |
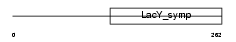
|
|||||
| Description | Vesicular acetylcholine transporter (VAChT) (Solute carrier family 18 member 3). | |||||
|
VACHT_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.069385 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16572 | Gene names | SLC18A3, VACHT | |||
|
Domain Architecture |
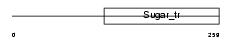
|
|||||
| Description | Vesicular acetylcholine transporter (VAChT) (Solute carrier family 18 member 3). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.062137 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.043058 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.041606 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.001096 (rank : 117) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.041865 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
EXTL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.015803 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKV7 | Gene names | Extl1 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 1 (EC 2.4.1.224) (Glucuronosyl-N-acetylglucosaminyl- proteoglycan 4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L) (Multiple exostosis-like protein). | |||||
|
IZUM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.034988 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D9J7 | Gene names | Izumo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Izumo sperm-egg fusion protein 1 precursor (Sperm-specific protein izumo). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.042981 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MIS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.021240 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27106 | Gene names | Amh | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
S23A2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.036498 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGH3, Q8WWR4, Q92512, Q96D54, Q9UNU1, Q9UP85 | Gene names | SLC23A2, KIAA0238, NBTL1, SVCT2, YSPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (hSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2) (Nucleobase transporter-like 1 protein). | |||||
|
S23A2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.036533 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPR4, Q8C327, Q9JM78 | Gene names | Slc23a2, Kiaa0238, Svct2, Yspl2 | |||
|
Domain Architecture |
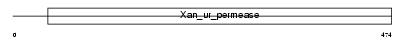
|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (mSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.025782 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
VMAT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.084919 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05940, Q15876, Q9H3P6 | Gene names | SLC18A2, SVMT, VMAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicular amine transporter (Monoamine transporter) (Vesicular amine transporter 2) (VAT2) (Solute carrier family 18 member 2). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.019139 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.011540 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.019809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.043933 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
VMAT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.088617 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BRU6, Q8CC55 | Gene names | Slc18a2, Vmat2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicular amine transporter (Monoamine transporter) (Vesicular amine transporter 2) (VAT2) (Solute carrier family 18 member 2). | |||||
|
COX11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.037649 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N1, Q9BRX0, Q9UME8 | Gene names | COX11 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome c oxidase assembly protein COX11, mitochondrial precursor. | |||||
|
LPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.019168 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.003013 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.019220 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
VMAT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.099130 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54219 | Gene names | SLC18A1, VMAT1 | |||
|
Domain Architecture |
|
|||||
| Description | Chromaffin granule amine transporter (Vesicular amine transporter 1) (VAT1) (Solute carrier family 18 member 1). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.008642 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.031647 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FLVC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.052531 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5Y0, Q86XY9, Q9NVR9 | Gene names | FLVCR1, FLVCR | |||
|
Domain Architecture |

|
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 1 (Feline leukemia virus subgroup C receptor) (hFLVCR). | |||||
|
COG8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.032492 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.007820 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
GTR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.009924 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11168 | Gene names | SLC2A2, GLUT2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 2 (Glucose transporter type 2, liver). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.015812 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.029256 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.012912 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.012636 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.027003 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009924 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.016310 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011013 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.013981 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GP137_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.015925 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14444, Q15074 | Gene names | M11S1, GPIP137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI-anchored protein p137 (p137GPI) (Membrane component chromosome 11 surface marker 1). | |||||
|
GP176_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.001387 (rank : 116) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.045721 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.004034 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.026452 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.025041 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ZN358_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.001025 (rank : 118) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
COFA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.014281 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.020790 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
FLVC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.047818 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPI3, Q96JY3, Q9NX90 | Gene names | FLVCR2, C14orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.013914 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.013841 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.038211 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.008934 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.058404 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
STAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.026522 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
TSSK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | -0.000652 (rank : 119) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BXA7 | Gene names | TSSK1, SPOGA1, SPOGA4, STK22A, STK22D | |||
|
Domain Architecture |
|
|||||
| Description | Testis-specific serine/threonine-protein kinase 1 (EC 2.7.11.1) (TSSK- 1) (Testis-specific kinase 1) (TSK-1) (Serine/threonine-protein kinase 22A). | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.001802 (rank : 115) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006815 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.060075 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.052377 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059500 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.052509 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.051356 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.051184 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052643 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.059338 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.063965 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.076351 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.067303 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.058745 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053729 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051194 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052228 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055859 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053448 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.054849 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051006 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.059978 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050738 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.063539 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
VMAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.081551 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R090 | Gene names | Slc18a1, Vmat1 | |||
|
Domain Architecture |
|
|||||
| Description | Chromaffin granule amine transporter (Vesicular amine transporter 1) (VAT1) (Solute carrier family 18 member 1). | |||||
|
MOT8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.984316 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MOT4_MOUSE
|
||||||
| NC score | 0.857556 (rank : 3) | θ value | 5.39604e-37 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P57787, Q9ES80, Q9ESF8 | Gene names | Slc16a3, Mct4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 4 (MCT 4) (Solute carrier family 16 member 3). | |||||
|
MOT4_HUMAN
|
||||||
| NC score | 0.857450 (rank : 4) | θ value | 7.04741e-37 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15427 | Gene names | SLC16A3, MCT3, MCT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 4 (MCT 4) (MCT 3) (Solute carrier family 16 member 3). | |||||
|
MOT3_MOUSE
|
||||||
| NC score | 0.856773 (rank : 5) | θ value | 9.20422e-37 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35308 | Gene names | Slc16a8, Mct3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 3 (MCT 3) (Solute carrier family 16 member 8) (Proton-coupled monocarboxylate transporter 3). | |||||
|
MOT3_HUMAN
|
||||||
| NC score | 0.852520 (rank : 6) | θ value | 2.77131e-33 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95907, Q9UBE2 | Gene names | SLC16A8, MCT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 3 (MCT 3) (Solute carrier family 16 member 8). | |||||
|
MOT2_HUMAN
|
||||||
| NC score | 0.849903 (rank : 7) | θ value | 3.16345e-37 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60669, Q9UPB3 | Gene names | SLC16A7, MCT2 | |||
|
Domain Architecture |
|
|||||
| Description | Monocarboxylate transporter 2 (MCT 2) (Solute carrier family 16 member 7). | |||||
|
MOT6_HUMAN
|
||||||
| NC score | 0.846490 (rank : 8) | θ value | 9.2256e-29 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15375 | Gene names | SLC16A5, MCT5, MCT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 6 (MCT 6) (MCT 5) (Solute carrier family 16 member 5). | |||||
|
MOT2_MOUSE
|
||||||
| NC score | 0.843236 (rank : 9) | θ value | 9.8567e-31 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70451 | Gene names | Slc16a7, Mct2 | |||
|
Domain Architecture |
|
|||||
| Description | Monocarboxylate transporter 2 (MCT 2) (Solute carrier family 16 member 7). | |||||
|
MOT1_HUMAN
|
||||||
| NC score | 0.841565 (rank : 10) | θ value | 2.34606e-32 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53985, Q9NSJ9 | Gene names | SLC16A1, MCT1 | |||
|
Domain Architecture |
|
|||||
| Description | Monocarboxylate transporter 1 (MCT 1) (Solute carrier family 16 member 1). | |||||
|
MOT1_MOUSE
|
||||||
| NC score | 0.838783 (rank : 11) | θ value | 4.89182e-30 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P53986 | Gene names | Slc16a1, Mct1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 1 (MCT 1) (Solute carrier family 16 member 1). | |||||
|
MOT7_HUMAN
|
||||||
| NC score | 0.799090 (rank : 12) | θ value | 2.7842e-17 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15403 | Gene names | SLC16A6, MCT6, MCT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 7 (MCT 7) (MCT 6) (Solute carrier family 16 member 6). | |||||
|
MOT5_HUMAN
|
||||||
| NC score | 0.779393 (rank : 13) | θ value | 1.52774e-15 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15374 | Gene names | SLC16A4, MCT4, MCT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 5 (MCT 5) (MCT 4) (Solute carrier family 16 member 4). | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.108352 (rank : 14) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
VMAT1_HUMAN
|
||||||
| NC score | 0.099130 (rank : 15) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54219 | Gene names | SLC18A1, VMAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromaffin granule amine transporter (Vesicular amine transporter 1) (VAT1) (Solute carrier family 18 member 1). | |||||
|
FLVC2_MOUSE
|
||||||
| NC score | 0.095020 (rank : 16) | θ value | 0.0148317 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91X85 | Gene names | Flvcr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
VMAT2_MOUSE
|
||||||
| NC score | 0.088617 (rank : 17) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BRU6, Q8CC55 | Gene names | Slc18a2, Vmat2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicular amine transporter (Monoamine transporter) (Vesicular amine transporter 2) (VAT2) (Solute carrier family 18 member 2). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.086100 (rank : 18) | θ value | 0.000602161 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
VMAT2_HUMAN
|
||||||
| NC score | 0.084919 (rank : 19) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05940, Q15876, Q9H3P6 | Gene names | SLC18A2, SVMT, VMAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicular amine transporter (Monoamine transporter) (Vesicular amine transporter 2) (VAT2) (Solute carrier family 18 member 2). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.082536 (rank : 20) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
VMAT1_MOUSE
|
||||||
| NC score | 0.081551 (rank : 21) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R090 | Gene names | Slc18a1, Vmat1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromaffin granule amine transporter (Vesicular amine transporter 1) (VAT1) (Solute carrier family 18 member 1). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.081523 (rank : 22) | θ value | 0.00298849 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.077053 (rank : 23) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.076351 (rank : 24) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
VACHT_MOUSE
|
||||||
| NC score | 0.075100 (rank : 25) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35304 | Gene names | Slc18a3, Vacht | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular acetylcholine transporter (VAChT) (Solute carrier family 18 member 3). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.073319 (rank : 26) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.071867 (rank : 27) | θ value | 0.000270298 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
VACHT_HUMAN
|
||||||
| NC score | 0.069385 (rank : 28) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16572 | Gene names | SLC18A3, VACHT | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular acetylcholine transporter (VAChT) (Solute carrier family 18 member 3). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.067303 (rank : 29) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.065195 (rank : 30) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.063965 (rank : 31) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.063539 (rank : 32) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.062137 (rank : 33) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.060075 (rank : 34) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.059978 (rank : 35) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.059813 (rank : 36) | θ value | 0.000461057 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.059500 (rank : 37) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.059338 (rank : 38) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.058825 (rank : 39) | θ value | 0.00134147 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.058745 (rank : 40) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.058404 (rank : 41) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.056813 (rank : 42) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SPR2D_HUMAN
|
||||||
| NC score | 0.055859 (rank : 43) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.055089 (rank : 44) | θ value | 0.000786445 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.054849 (rank : 45) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.053729 (rank : 46) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.053448 (rank : 47) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.052643 (rank : 48) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
FLVC1_HUMAN
|
||||||
| NC score | 0.052531 (rank : 49) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5Y0, Q86XY9, Q9NVR9 | Gene names | FLVCR1, FLVCR | |||
|
Domain Architecture |

|
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 1 (Feline leukemia virus subgroup C receptor) (hFLVCR). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.052509 (rank : 50) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.052377 (rank : 51) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SPR2B_HUMAN
|
||||||
| NC score | 0.052228 (rank : 52) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.051356 (rank : 53) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.051194 (rank : 54) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.051184 (rank : 55) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.051006 (rank : 56) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.050738 (rank : 57) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.048303 (rank : 58) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.048220 (rank : 59) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
FLVC2_HUMAN
|
||||||
| NC score | 0.047818 (rank : 60) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPI3, Q96JY3, Q9NX90 | Gene names | FLVCR2, C14orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.045803 (rank : 61) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.045721 (rank : 62) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.043933 (rank : 63) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.043058 (rank : 64) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.042981 (rank : 65) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.041865 (rank : 66) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.041606 (rank : 67) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.039953 (rank : 68) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.038211 (rank : 69) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.037902 (rank : 70) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
COX11_HUMAN
|
||||||
| NC score | 0.037649 (rank : 71) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N1, Q9BRX0, Q9UME8 | Gene names | COX11 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c oxidase assembly protein COX11, mitochondrial precursor. | |||||
|
S17A5_HUMAN
|
||||||
| NC score | 0.037532 (rank : 72) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRA2, Q8NBR5, Q9UGH0 | Gene names | SLC17A5 | |||
|
Domain Architecture |

|
|||||
| Description | Sialin (Solute carrier family 17 member 5) (Sodium/sialic acid cotransporter) (AST) (Membrane glycoprotein HP59). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.036704 (rank : 73) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
S23A2_MOUSE
|
||||||
| NC score | 0.036533 (rank : 74) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPR4, Q8C327, Q9JM78 | Gene names | Slc23a2, Kiaa0238, Svct2, Yspl2 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (mSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2). | |||||
|
S23A2_HUMAN
|
||||||
| NC score | 0.036498 (rank : 75) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGH3, Q8WWR4, Q92512, Q96D54, Q9UNU1, Q9UP85 | Gene names | SLC23A2, KIAA0238, NBTL1, SVCT2, YSPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (hSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2) (Nucleobase transporter-like 1 protein). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.036077 (rank : 76) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
IZUM1_MOUSE
|
||||||
| NC score | 0.034988 (rank : 77) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D9J7 | Gene names | Izumo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Izumo sperm-egg fusion protein 1 precursor (Sperm-specific protein izumo). | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.032492 (rank : 78) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.031647 (rank : 79) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.029256 (rank : 80) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.027003 (rank : 81) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
STAT2_HUMAN
|
||||||
| NC score | 0.026522 (rank : 82) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52630, Q16430, Q16431 | Gene names | STAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2 (p113). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.026452 (rank : 83) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.026277 (rank : 84) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
SRC8_MOUSE
|
||||||
| NC score | 0.025838 (rank : 85) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
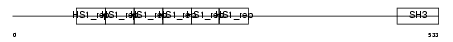
|
|||||
| Description | Src substrate cortactin. | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.025782 (rank : 86) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.025041 (rank : 87) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.024546 (rank : 88) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
MIS_MOUSE
|
||||||
| NC score | 0.021240 (rank : 89) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27106 | Gene names | Amh | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.020790 (rank : 90) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.019809 (rank : 91) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.019220 (rank : 92) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.019168 (rank : 93) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.019139 (rank : 94) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.016310 (rank : 95) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
GP137_HUMAN
|
||||||
| NC score | 0.015925 (rank : 96) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14444, Q15074 | Gene names | M11S1, GPIP137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI-anchored protein p137 (p137GPI) (Membrane component chromosome 11 surface marker 1). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.015812 (rank : 97) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
EXTL1_MOUSE
|
||||||
| NC score | 0.015803 (rank : 98) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKV7 | Gene names | Extl1 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 1 (EC 2.4.1.224) (Glucuronosyl-N-acetylglucosaminyl- proteoglycan 4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L) (Multiple exostosis-like protein). | |||||
|
COFA1_MOUSE
|
||||||
| NC score | 0.014281 (rank : 99) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.013981 (rank : 100) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.013914 (rank : 101) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.013841 (rank : 102) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.012912 (rank : 103) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.012636 (rank : 104) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.011540 (rank : 105) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.011013 (rank : 106) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.009924 (rank : 107) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
GTR2_HUMAN
|
||||||
| NC score | 0.009924 (rank : 108) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11168 | Gene names | SLC2A2, GLUT2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 2 (Glucose transporter type 2, liver). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.008934 (rank : 109) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.008642 (rank : 110) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.007820 (rank : 111) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.006815 (rank : 112) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.004034 (rank : 113) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.003013 (rank : 114) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.001802 (rank : 115) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
GP176_MOUSE
|
||||||
| NC score | 0.001387 (rank : 116) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.001096 (rank : 117) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ZN358_HUMAN
|
||||||
| NC score | 0.001025 (rank : 118) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
TSSK1_HUMAN
|
||||||
| NC score | -0.000652 (rank : 119) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BXA7 | Gene names | TSSK1, SPOGA1, SPOGA4, STK22A, STK22D | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific serine/threonine-protein kinase 1 (EC 2.7.11.1) (TSSK- 1) (Testis-specific kinase 1) (TSK-1) (Serine/threonine-protein kinase 22A). | |||||