Please be patient as the page loads
|
AKAP3_MOUSE
|
||||||
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AKAP3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989278 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75969, O75945, Q9UM61 | Gene names | AKAP3, AKAP110, SOB1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte- binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95). | |||||
|
AKAP3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
AKAP4_MOUSE
|
||||||
| θ value | 1.64275e-110 (rank : 3) | NC score | 0.915086 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60662, Q60630, Q8R2G7 | Gene names | Akap4, Akap82, Fsc1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (mAKAP82) (Major sperm fibrous sheath protein) (FSC1) (p82). | |||||
|
AKAP4_HUMAN
|
||||||
| θ value | 5.11853e-104 (rank : 4) | NC score | 0.914404 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JQC9, O60904, O95246, Q5JQD1, Q5JQD2, Q5JQD3 | Gene names | AKAP4, AKAP82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (hAKAP82) (Major sperm fibrous sheath protein) (HI). | |||||
|
AKA11_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 5) | NC score | 0.336716 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKA4, O75124, Q9NUK7 | Gene names | AKAP11, AKAP220, KIAA0629 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 11 (Protein kinase A-anchoring protein 11) (PRKA11) (A kinase anchor protein 220 kDa) (AKAP 220) (hAKAP220). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.054585 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.030859 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
BTG3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.039901 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50615, Q62328 | Gene names | Btg3, Ana, Tob5 | |||
|
Domain Architecture |

|
|||||
| Description | BTG3 protein (Tob5 protein) (Abundant in neuroepithelium area protein). | |||||
|
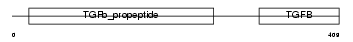
TGFB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.023282 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27090 | Gene names | Tgfb2 | |||
|
Domain Architecture |
|
|||||
| Description | Transforming growth factor beta-2 precursor (TGF-beta-2). | |||||
|
PARG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.058896 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PARG_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.058968 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
BTG3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.034092 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14201, Q96ET7 | Gene names | BTG3, ANA, TOB5 | |||
|
Domain Architecture |

|
|||||
| Description | BTG3 protein (Tob5 protein) (Abundant in neuroepithelium area protein). | |||||
|
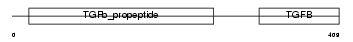
TGFB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.019787 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61812, P08112, Q15579, Q15581, Q4VAV9 | Gene names | TGFB2 | |||
|
Domain Architecture |
|
|||||
| Description | Transforming growth factor beta-2 precursor (TGF-beta-2) (Glioblastoma-derived T-cell suppressor factor) (G-TSF) (BSC-1 cell growth inhibitor) (Polyergin) (Cetermin). | |||||
|
K0056_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.018564 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.007698 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
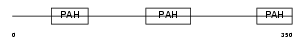
SIN3B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.014555 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62141, O54976, Q6A013, Q8VCB8, Q8VDZ5 | Gene names | Sin3b, Kiaa0700 | |||
|
Domain Architecture |
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.003135 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
DCTN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.018580 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJW0, Q8TAN8 | Gene names | DCTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
DCTN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.018580 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBY8, Q923A0, Q9D4X0 | Gene names | Dctn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
RAI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.012763 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
SQSTM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.011908 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
TDP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.010141 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08639 | Gene names | Tfdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-1 (E2F dimerization partner 1) (DRTF1- polypeptide 1). | |||||
|
AKAP3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
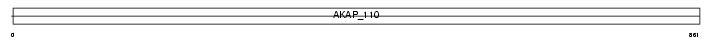
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
AKAP3_HUMAN
|
||||||
| NC score | 0.989278 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75969, O75945, Q9UM61 | Gene names | AKAP3, AKAP110, SOB1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte- binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95). | |||||
|
AKAP4_MOUSE
|
||||||
| NC score | 0.915086 (rank : 3) | θ value | 1.64275e-110 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60662, Q60630, Q8R2G7 | Gene names | Akap4, Akap82, Fsc1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (mAKAP82) (Major sperm fibrous sheath protein) (FSC1) (p82). | |||||
|
AKAP4_HUMAN
|
||||||
| NC score | 0.914404 (rank : 4) | θ value | 5.11853e-104 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JQC9, O60904, O95246, Q5JQD1, Q5JQD2, Q5JQD3 | Gene names | AKAP4, AKAP82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (hAKAP82) (Major sperm fibrous sheath protein) (HI). | |||||
|
AKA11_HUMAN
|
||||||
| NC score | 0.336716 (rank : 5) | θ value | 5.81887e-07 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKA4, O75124, Q9NUK7 | Gene names | AKAP11, AKAP220, KIAA0629 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 11 (Protein kinase A-anchoring protein 11) (PRKA11) (A kinase anchor protein 220 kDa) (AKAP 220) (hAKAP220). | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.058968 (rank : 6) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.058896 (rank : 7) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.054585 (rank : 8) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
BTG3_MOUSE
|
||||||
| NC score | 0.039901 (rank : 9) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50615, Q62328 | Gene names | Btg3, Ana, Tob5 | |||
|
Domain Architecture |

|
|||||
| Description | BTG3 protein (Tob5 protein) (Abundant in neuroepithelium area protein). | |||||
|
BTG3_HUMAN
|
||||||
| NC score | 0.034092 (rank : 10) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14201, Q96ET7 | Gene names | BTG3, ANA, TOB5 | |||
|
Domain Architecture |

|
|||||
| Description | BTG3 protein (Tob5 protein) (Abundant in neuroepithelium area protein). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.030859 (rank : 11) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
TGFB2_MOUSE
|
||||||
| NC score | 0.023282 (rank : 12) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27090 | Gene names | Tgfb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor beta-2 precursor (TGF-beta-2). | |||||
|
TGFB2_HUMAN
|
||||||
| NC score | 0.019787 (rank : 13) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61812, P08112, Q15579, Q15581, Q4VAV9 | Gene names | TGFB2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor beta-2 precursor (TGF-beta-2) (Glioblastoma-derived T-cell suppressor factor) (G-TSF) (BSC-1 cell growth inhibitor) (Polyergin) (Cetermin). | |||||
|
DCTN4_HUMAN
|
||||||
| NC score | 0.018580 (rank : 14) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJW0, Q8TAN8 | Gene names | DCTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
DCTN4_MOUSE
|
||||||
| NC score | 0.018580 (rank : 15) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBY8, Q923A0, Q9D4X0 | Gene names | Dctn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynactin subunit 4 (Dynactin subunit p62). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.018564 (rank : 16) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
SIN3B_MOUSE
|
||||||
| NC score | 0.014555 (rank : 17) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62141, O54976, Q6A013, Q8VCB8, Q8VDZ5 | Gene names | Sin3b, Kiaa0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
RAI2_HUMAN
|
||||||
| NC score | 0.012763 (rank : 18) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
SQSTM_MOUSE
|
||||||
| NC score | 0.011908 (rank : 19) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
TDP1_MOUSE
|
||||||
| NC score | 0.010141 (rank : 20) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08639 | Gene names | Tfdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-1 (E2F dimerization partner 1) (DRTF1- polypeptide 1). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.007698 (rank : 21) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.003135 (rank : 22) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||