Please be patient as the page loads
|
K0355_HUMAN
|
||||||
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0355_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 2) | NC score | 0.105201 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 3) | NC score | 0.076352 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 4) | NC score | 0.092910 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ACCN4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.090273 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
ACCN4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.088545 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.081723 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.082206 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.053572 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.063176 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.016093 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
LIPB2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.051854 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.051255 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.043653 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.040924 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CTF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.067394 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60753 | Gene names | Ctf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cardiotrophin-1 (CT-1). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.054732 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.048177 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.048274 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.037357 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
TMM24_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.049510 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.038758 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
KIF1C_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.016488 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.038103 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.019251 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.040085 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ROBO2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.015254 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
ZN692_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.009245 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.020187 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.026813 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.034069 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.021023 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.026678 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.039307 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
NT3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.031909 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20181 | Gene names | Ntf3, Ntf-3 | |||
|
Domain Architecture |
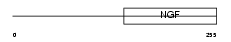
|
|||||
| Description | Neurotrophin-3 precursor (NT-3) (Neurotrophic factor) (HDNF) (Nerve growth factor 2) (NGF-2). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.065905 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.013784 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
PAK7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.001494 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P286, Q5W115, Q9BX09, Q9ULF6 | Gene names | PAK7, KIAA1264, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (PAK-5). | |||||
|
PHF8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.015919 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.041183 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.030101 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.024612 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.008836 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.027856 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.050611 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.036710 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
ACCN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.053861 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.053747 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.026156 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
G3ST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.016848 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RP7, Q8N3P7, Q8WZ17, Q96E33, Q9HA78 | Gene names | GAL3ST4 | |||
|
Domain Architecture |
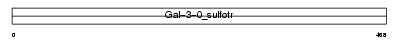
|
|||||
| Description | Galactose-3-O-sulfotransferase 4 (EC 2.8.2.-) (Galbeta1-3GalNAc 3'- sulfotransferase) (Beta-galactose-3-O-sulfotransferase 4). | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.005873 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
NT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.025588 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20783 | Gene names | NTF3 | |||
|
Domain Architecture |
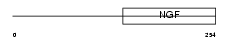
|
|||||
| Description | Neurotrophin-3 precursor (NT-3) (Neurotrophic factor) (HDNF) (Nerve growth factor 2) (NGF-2). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.031215 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
STML1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.015732 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBI4, O95675, Q6FGL8, Q8WYI7, Q9UMB9, Q9UMC0, Q9Y6H9 | Gene names | STOML1, SLP1, UNC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1) (Stomatin-related protein) (STORP) (EPB72-like 1) (UNC24 homolog). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.038344 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.066512 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.061285 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.055259 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.061296 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.059627 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.066924 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
K0355_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.105201 (rank : 2) | θ value | 0.000461057 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.092910 (rank : 3) | θ value | 0.0330416 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ACCN4_HUMAN
|
||||||
| NC score | 0.090273 (rank : 4) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
ACCN4_MOUSE
|
||||||
| NC score | 0.088545 (rank : 5) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.082206 (rank : 6) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.081723 (rank : 7) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.076352 (rank : 8) | θ value | 0.0193708 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CTF1_MOUSE
|
||||||
| NC score | 0.067394 (rank : 9) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60753 | Gene names | Ctf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cardiotrophin-1 (CT-1). | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.066924 (rank : 10) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.066512 (rank : 11) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.065905 (rank : 12) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.063176 (rank : 13) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.061296 (rank : 14) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.061285 (rank : 15) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.059627 (rank : 16) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.055259 (rank : 17) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.054732 (rank : 18) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ACCN2_HUMAN
|
||||||
| NC score | 0.053861 (rank : 19) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN2_MOUSE
|
||||||
| NC score | 0.053747 (rank : 20) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.053572 (rank : 21) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
LIPB2_HUMAN
|
||||||
| NC score | 0.051854 (rank : 22) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.051255 (rank : 23) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.050611 (rank : 24) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
TMM24_MOUSE
|
||||||
| NC score | 0.049510 (rank : 25) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.048274 (rank : 26) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.048177 (rank : 27) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.043653 (rank : 28) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.041183 (rank : 29) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.040924 (rank : 30) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.040085 (rank : 31) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.039307 (rank : 32) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.038758 (rank : 33) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.038344 (rank : 34) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.038103 (rank : 35) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.037357 (rank : 36) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.036710 (rank : 37) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.034069 (rank : 38) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
NT3_MOUSE
|
||||||
| NC score | 0.031909 (rank : 39) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20181 | Gene names | Ntf3, Ntf-3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrophin-3 precursor (NT-3) (Neurotrophic factor) (HDNF) (Nerve growth factor 2) (NGF-2). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.031215 (rank : 40) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.030101 (rank : 41) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.027856 (rank : 42) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.026813 (rank : 43) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.026678 (rank : 44) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.026156 (rank : 45) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
NT3_HUMAN
|
||||||
| NC score | 0.025588 (rank : 46) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20783 | Gene names | NTF3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrophin-3 precursor (NT-3) (Neurotrophic factor) (HDNF) (Nerve growth factor 2) (NGF-2). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.024612 (rank : 47) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.021023 (rank : 48) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.020187 (rank : 49) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.019251 (rank : 50) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
G3ST4_HUMAN
|
||||||
| NC score | 0.016848 (rank : 51) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RP7, Q8N3P7, Q8WZ17, Q96E33, Q9HA78 | Gene names | GAL3ST4 | |||
|
Domain Architecture |
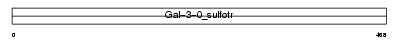
|
|||||
| Description | Galactose-3-O-sulfotransferase 4 (EC 2.8.2.-) (Galbeta1-3GalNAc 3'- sulfotransferase) (Beta-galactose-3-O-sulfotransferase 4). | |||||
|
KIF1C_MOUSE
|
||||||
| NC score | 0.016488 (rank : 52) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.016093 (rank : 53) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.015919 (rank : 54) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
STML1_HUMAN
|
||||||
| NC score | 0.015732 (rank : 55) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBI4, O95675, Q6FGL8, Q8WYI7, Q9UMB9, Q9UMC0, Q9Y6H9 | Gene names | STOML1, SLP1, UNC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1) (Stomatin-related protein) (STORP) (EPB72-like 1) (UNC24 homolog). | |||||
|
ROBO2_HUMAN
|
||||||
| NC score | 0.015254 (rank : 56) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.013784 (rank : 57) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
ZN692_MOUSE
|
||||||
| NC score | 0.009245 (rank : 58) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.008836 (rank : 59) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
KLF17_HUMAN
|
||||||
| NC score | 0.005873 (rank : 60) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
PAK7_HUMAN
|
||||||
| NC score | 0.001494 (rank : 61) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P286, Q5W115, Q9BX09, Q9ULF6 | Gene names | PAK7, KIAA1264, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (PAK-5). | |||||