Please be patient as the page loads
|
ACCN2_MOUSE
|
||||||
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACCN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996775 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16515, Q13553, Q8N3E2 | Gene names | ACCN1, ACCN, BNAC1, MDEG | |||
|
Domain Architecture |
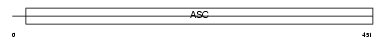
|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Amiloride-sensitive cation channel neuronal 1) (Acid-sensing ion channel 2) (ASIC2) (Brain sodium channel 1) (BNaC1) (BNC1) (Mammalian degenerin homolog). | |||||
|
ACCN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996780 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q925H0, Q5SUU2, Q61203 | Gene names | Accn1, Asic2, Bnac1 | |||
|
Domain Architecture |
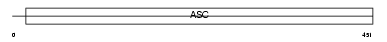
|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Acid-sensing ion channel 2) (Brain sodium channel 1) (BNaC1). | |||||
|
ACCN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999782 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ACCN3_MOUSE
|
||||||
| θ value | 1.79534e-149 (rank : 5) | NC score | 0.988824 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6X1Y6, Q7TQH4 | Gene names | Accn3, Asic3, Drasic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (Dorsal root ASIC) (DRASIC). | |||||
|
ACCN3_HUMAN
|
||||||
| θ value | 7.30591e-143 (rank : 6) | NC score | 0.988008 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UHC3, O60263, O75906, Q59FN9, Q9UER8, Q9UHC4 | Gene names | ACCN3, ASIC3, SLNAC1, TNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (hASIC3) (Testis sodium channel 1) (hTNaC1). | |||||
|
ACCN4_MOUSE
|
||||||
| θ value | 1.33454e-136 (rank : 7) | NC score | 0.985939 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ACCN4_HUMAN
|
||||||
| θ value | 1.74701e-128 (rank : 8) | NC score | 0.982125 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
SCNNB_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 9) | NC score | 0.802661 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51168, O60891, Q96KG2, Q9UJ32, Q9UMU5 | Gene names | SCNN1B | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH) (ENaCB). | |||||
|
SCNNB_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 10) | NC score | 0.793278 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU38 | Gene names | Scnn1b | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH). | |||||
|
SCNNA_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 11) | NC score | 0.789164 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNA_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 12) | NC score | 0.781662 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |
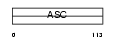
|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNG_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 13) | NC score | 0.775964 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU39 | Gene names | Scnn1g | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNG_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 14) | NC score | 0.761181 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51170, P78437, Q6PCC2, Q93023, Q93024, Q93025, Q93026, Q93027, Q96TD2 | Gene names | SCNN1G | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNND_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 15) | NC score | 0.783756 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.013697 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.014107 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.004053 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
ATS20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.002503 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
K0355_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.053747 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
ACCN2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ACCN2_HUMAN
|
||||||
| NC score | 0.999782 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN1_MOUSE
|
||||||
| NC score | 0.996780 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q925H0, Q5SUU2, Q61203 | Gene names | Accn1, Asic2, Bnac1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Acid-sensing ion channel 2) (Brain sodium channel 1) (BNaC1). | |||||
|
ACCN1_HUMAN
|
||||||
| NC score | 0.996775 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16515, Q13553, Q8N3E2 | Gene names | ACCN1, ACCN, BNAC1, MDEG | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Amiloride-sensitive cation channel neuronal 1) (Acid-sensing ion channel 2) (ASIC2) (Brain sodium channel 1) (BNaC1) (BNC1) (Mammalian degenerin homolog). | |||||
|
ACCN3_MOUSE
|
||||||
| NC score | 0.988824 (rank : 5) | θ value | 1.79534e-149 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6X1Y6, Q7TQH4 | Gene names | Accn3, Asic3, Drasic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (Dorsal root ASIC) (DRASIC). | |||||
|
ACCN3_HUMAN
|
||||||
| NC score | 0.988008 (rank : 6) | θ value | 7.30591e-143 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UHC3, O60263, O75906, Q59FN9, Q9UER8, Q9UHC4 | Gene names | ACCN3, ASIC3, SLNAC1, TNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (hASIC3) (Testis sodium channel 1) (hTNaC1). | |||||
|
ACCN4_MOUSE
|
||||||
| NC score | 0.985939 (rank : 7) | θ value | 1.33454e-136 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ACCN4_HUMAN
|
||||||
| NC score | 0.982125 (rank : 8) | θ value | 1.74701e-128 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
SCNNB_HUMAN
|
||||||
| NC score | 0.802661 (rank : 9) | θ value | 5.584e-34 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51168, O60891, Q96KG2, Q9UJ32, Q9UMU5 | Gene names | SCNN1B | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH) (ENaCB). | |||||
|
SCNNB_MOUSE
|
||||||
| NC score | 0.793278 (rank : 10) | θ value | 1.98606e-31 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU38 | Gene names | Scnn1b | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH). | |||||
|
SCNNA_HUMAN
|
||||||
| NC score | 0.789164 (rank : 11) | θ value | 3.74554e-30 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNND_HUMAN
|
||||||
| NC score | 0.783756 (rank : 12) | θ value | 2.12685e-25 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
SCNNA_MOUSE
|
||||||
| NC score | 0.781662 (rank : 13) | θ value | 9.2256e-29 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNG_MOUSE
|
||||||
| NC score | 0.775964 (rank : 14) | θ value | 9.2256e-29 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU39 | Gene names | Scnn1g | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNG_HUMAN
|
||||||
| NC score | 0.761181 (rank : 15) | θ value | 8.63488e-27 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51170, P78437, Q6PCC2, Q93023, Q93024, Q93025, Q93026, Q93027, Q96TD2 | Gene names | SCNN1G | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
K0355_HUMAN
|
||||||
| NC score | 0.053747 (rank : 16) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.014107 (rank : 17) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.013697 (rank : 18) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.004053 (rank : 19) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
ATS20_HUMAN
|
||||||
| NC score | 0.002503 (rank : 20) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||