Please be patient as the page loads
|
SCNNA_MOUSE
|
||||||
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |
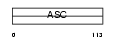
|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SCNNA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997673 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNND_HUMAN
|
||||||
| θ value | 2.52853e-119 (rank : 3) | NC score | 0.976690 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
SCNNG_HUMAN
|
||||||
| θ value | 3.00075e-104 (rank : 4) | NC score | 0.962981 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51170, P78437, Q6PCC2, Q93023, Q93024, Q93025, Q93026, Q93027, Q96TD2 | Gene names | SCNN1G | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNG_MOUSE
|
||||||
| θ value | 3.91911e-104 (rank : 5) | NC score | 0.964585 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU39 | Gene names | Scnn1g | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNB_HUMAN
|
||||||
| θ value | 3.00772e-96 (rank : 6) | NC score | 0.957195 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51168, O60891, Q96KG2, Q9UJ32, Q9UMU5 | Gene names | SCNN1B | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH) (ENaCB). | |||||
|
SCNNB_MOUSE
|
||||||
| θ value | 6.27148e-94 (rank : 7) | NC score | 0.957387 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU38 | Gene names | Scnn1b | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH). | |||||
|
ACCN3_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 8) | NC score | 0.781684 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UHC3, O60263, O75906, Q59FN9, Q9UER8, Q9UHC4 | Gene names | ACCN3, ASIC3, SLNAC1, TNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (hASIC3) (Testis sodium channel 1) (hTNaC1). | |||||
|
ACCN1_HUMAN
|
||||||
| θ value | 9.2256e-29 (rank : 9) | NC score | 0.788433 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16515, Q13553, Q8N3E2 | Gene names | ACCN1, ACCN, BNAC1, MDEG | |||
|
Domain Architecture |
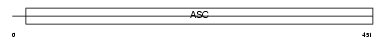
|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Amiloride-sensitive cation channel neuronal 1) (Acid-sensing ion channel 2) (ASIC2) (Brain sodium channel 1) (BNaC1) (BNC1) (Mammalian degenerin homolog). | |||||
|
ACCN1_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 10) | NC score | 0.788091 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q925H0, Q5SUU2, Q61203 | Gene names | Accn1, Asic2, Bnac1 | |||
|
Domain Architecture |
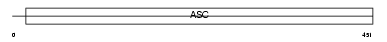
|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Acid-sensing ion channel 2) (Brain sodium channel 1) (BNaC1). | |||||
|
ACCN2_MOUSE
|
||||||
| θ value | 9.2256e-29 (rank : 11) | NC score | 0.781662 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ACCN2_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 12) | NC score | 0.778319 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN3_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 13) | NC score | 0.774998 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6X1Y6, Q7TQH4 | Gene names | Accn3, Asic3, Drasic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (Dorsal root ASIC) (DRASIC). | |||||
|
ACCN4_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 14) | NC score | 0.782592 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ACCN4_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 15) | NC score | 0.776214 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.023602 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
SRPK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.005118 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.012706 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.008074 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.015491 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.014319 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
SH22A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.012497 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.004249 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
KIRR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.003575 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TSU7, Q7TQ98 | Gene names | Kirrel2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 2 precursor (Kin of irregular chiasm-like protein 2). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.006140 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.008216 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
SL9A5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.006591 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
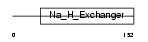
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.008669 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
RAD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.008512 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61550, P70219, Q91VB9, Q9DBU4 | Gene names | Rad21, Hr21 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand-break repair protein rad21 homolog (Pokeweed agglutinin- binding protein 29) (PW29) (SCC1 homolog). | |||||
|
SCNNA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNA_HUMAN
|
||||||
| NC score | 0.997673 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNND_HUMAN
|
||||||
| NC score | 0.976690 (rank : 3) | θ value | 2.52853e-119 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
SCNNG_MOUSE
|
||||||
| NC score | 0.964585 (rank : 4) | θ value | 3.91911e-104 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU39 | Gene names | Scnn1g | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNG_HUMAN
|
||||||
| NC score | 0.962981 (rank : 5) | θ value | 3.00075e-104 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51170, P78437, Q6PCC2, Q93023, Q93024, Q93025, Q93026, Q93027, Q96TD2 | Gene names | SCNN1G | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNB_MOUSE
|
||||||
| NC score | 0.957387 (rank : 6) | θ value | 6.27148e-94 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU38 | Gene names | Scnn1b | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH). | |||||
|
SCNNB_HUMAN
|
||||||
| NC score | 0.957195 (rank : 7) | θ value | 3.00772e-96 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51168, O60891, Q96KG2, Q9UJ32, Q9UMU5 | Gene names | SCNN1B | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH) (ENaCB). | |||||
|
ACCN1_HUMAN
|
||||||
| NC score | 0.788433 (rank : 8) | θ value | 9.2256e-29 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16515, Q13553, Q8N3E2 | Gene names | ACCN1, ACCN, BNAC1, MDEG | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Amiloride-sensitive cation channel neuronal 1) (Acid-sensing ion channel 2) (ASIC2) (Brain sodium channel 1) (BNaC1) (BNC1) (Mammalian degenerin homolog). | |||||
|
ACCN1_MOUSE
|
||||||
| NC score | 0.788091 (rank : 9) | θ value | 9.2256e-29 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q925H0, Q5SUU2, Q61203 | Gene names | Accn1, Asic2, Bnac1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Acid-sensing ion channel 2) (Brain sodium channel 1) (BNaC1). | |||||
|
ACCN4_MOUSE
|
||||||
| NC score | 0.782592 (rank : 10) | θ value | 8.63488e-27 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ACCN3_HUMAN
|
||||||
| NC score | 0.781684 (rank : 11) | θ value | 2.19584e-30 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UHC3, O60263, O75906, Q59FN9, Q9UER8, Q9UHC4 | Gene names | ACCN3, ASIC3, SLNAC1, TNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (hASIC3) (Testis sodium channel 1) (hTNaC1). | |||||
|
ACCN2_MOUSE
|
||||||
| NC score | 0.781662 (rank : 12) | θ value | 9.2256e-29 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ACCN2_HUMAN
|
||||||
| NC score | 0.778319 (rank : 13) | θ value | 7.80994e-28 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN4_HUMAN
|
||||||
| NC score | 0.776214 (rank : 14) | θ value | 7.30988e-26 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
ACCN3_MOUSE
|
||||||
| NC score | 0.774998 (rank : 15) | θ value | 1.73987e-27 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6X1Y6, Q7TQH4 | Gene names | Accn3, Asic3, Drasic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (Dorsal root ASIC) (DRASIC). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.023602 (rank : 16) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.015491 (rank : 17) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.014319 (rank : 18) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.012706 (rank : 19) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
SH22A_HUMAN
|
||||||
| NC score | 0.012497 (rank : 20) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.008669 (rank : 21) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
RAD21_MOUSE
|
||||||
| NC score | 0.008512 (rank : 22) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61550, P70219, Q91VB9, Q9DBU4 | Gene names | Rad21, Hr21 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand-break repair protein rad21 homolog (Pokeweed agglutinin- binding protein 29) (PW29) (SCC1 homolog). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.008216 (rank : 23) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.008074 (rank : 24) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SL9A5_HUMAN
|
||||||
| NC score | 0.006591 (rank : 25) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.006140 (rank : 26) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRPK1_HUMAN
|
||||||
| NC score | 0.005118 (rank : 27) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.004249 (rank : 28) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
KIRR2_MOUSE
|
||||||
| NC score | 0.003575 (rank : 29) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TSU7, Q7TQ98 | Gene names | Kirrel2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 2 precursor (Kin of irregular chiasm-like protein 2). | |||||