Please be patient as the page loads
|
TOIP2_HUMAN
|
||||||
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TOIP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 6.80647e-157 (rank : 2) | NC score | 0.926659 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TOIP1_HUMAN
|
||||||
| θ value | 7.45998e-71 (rank : 3) | NC score | 0.886520 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5JTV8, Q5JTV6, Q8IZ65, Q9H8Y6, Q9HAJ1, Q9NV52, Q9Y3X5 | Gene names | TOR1AIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1. | |||||
|
TOIP1_MOUSE
|
||||||
| θ value | 2.24614e-67 (rank : 4) | NC score | 0.887408 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921T2, Q3U7A4 | Gene names | Tor1aip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1 (Lamina-associated polypeptide 1B). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 5) | NC score | 0.120229 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.155398 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.089126 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 8) | NC score | 0.128550 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.095678 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.117918 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.152478 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.067539 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.042352 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.093539 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.093627 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.129408 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SH3BG_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.078372 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.035978 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.014236 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.060389 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
TR10A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.038056 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |
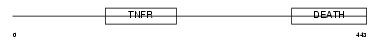
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
GZF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.002502 (rank : 118) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H116, Q96N08, Q9BQK9, Q9H117, Q9H6W6 | Gene names | GZF1, ZBTB23, ZNF336 | |||
|
Domain Architecture |
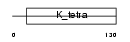
|
|||||
| Description | GDNF-inducible zinc finger protein 1 (Zinc finger protein 336) (Zinc finger and BTB domain-containing protein 23). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.063029 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.063245 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SHD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.035146 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
ANR15_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.019538 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.143631 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.035919 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.022504 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.054941 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.064651 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.068221 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
IF39_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.064857 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.044170 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.048934 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.014332 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
HERPU_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.040625 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJK5 | Gene names | Herpud1 | |||
|
Domain Architecture |

|
|||||
| Description | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein. | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.030966 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
ARS2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.065348 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.087096 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
I28RA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.035326 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IU57, Q6ZML8, Q8IZI7, Q8IZI8 | Gene names | IL28RA, IFNLR1, LICR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-28 receptor alpha chain precursor (IL-28R-alpha) (IL-28RA) (Cytokine receptor family 2 member 12) (Cytokine receptor class-II member 12) (CRF2-12) (Interferon lambda receptor 1) (IFN-lambda R1) (Likely interleukin or cytokine receptor 2). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.059214 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.020688 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.038160 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
S30BP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.040537 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHR5, Q8N1R5, Q8TDR8, Q96D27 | Gene names | SAP30BP, HCNGP, HTRG, HTRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.014159 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.038431 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.060542 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.044264 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.036236 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.024823 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.050702 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.010161 (rank : 107) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LMBRL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.024993 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX01, Q969J4, Q96BY8, Q96HN8, Q9NT09, Q9NVE1, Q9NVU9, Q9ULP6 | Gene names | LMBR1L, KIAA1174, LIMR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LMBR1L (Lipocalin-1-interacting membrane receptor) (Lipocalin- interacting membrane receptor) (Limb region 1 protein homolog-like). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.006759 (rank : 112) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
RPA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.023109 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.051199 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.056829 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.045640 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.032428 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.066673 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
LRCH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.008119 (rank : 108) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62046 | Gene names | Lrch1, Chdc1, Kiaa1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.010343 (rank : 106) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.005421 (rank : 114) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.068957 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.003822 (rank : 116) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.030287 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
BRMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.022184 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCU9 | Gene names | BRMS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer metastasis-suppressor 1. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.018752 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.054118 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.023576 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PSMD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.019259 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14685, Q99LL7 | Gene names | Psmd3, P91a, Tstap91a | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58) (Transplantation antigen P91A) (Tum-P91A antigen). | |||||
|
SH3BG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.036764 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55822, Q9BRB8 | Gene names | SH3BGR | |||
|
Domain Architecture |
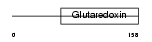
|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein) (21- glutamic acid-rich protein) (21-GARP). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.007961 (rank : 109) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.010968 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TNR16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.015057 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
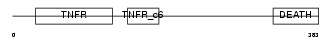
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.022848 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DBR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.020811 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923B1, Q8C1T9, Q8C7J7, Q99MT1 | Gene names | Dbr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lariat debranching enzyme (EC 3.1.-.-). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.010419 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
IP3KC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.013697 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.094131 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PARG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.029729 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.034444 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.010461 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
TF2AA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.019477 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
TIM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.015127 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1X4, Q63ZX9, Q6P204, Q6PDL4, Q7TPV8, Q8R0Q2, Q9R268, Q9Z0E7 | Gene names | Timeless, Tim1, Timeless1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (mTim). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.004862 (rank : 115) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.017025 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.007869 (rank : 110) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.011561 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.019148 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.006324 (rank : 113) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.007473 (rank : 111) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
LMBRL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.020212 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1E5, Q3TD82, Q69ZP7 | Gene names | Lmbr1l, D15Ertd735e, Kiaa1174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LMBR1L (Uteroglobin receptor). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.000945 (rank : 119) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.020937 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.021311 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
PCDG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.000779 (rank : 120) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.023484 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SG16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.052635 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
SSFA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.016967 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28290, Q96E36 | Gene names | SSFA2, CS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-specific antigen 2 (Cleavage signal-1 protein) (CS-1). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.002834 (rank : 117) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.062879 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.056076 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053368 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.072910 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.068855 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.052374 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.051424 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.057418 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.053696 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.051092 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.057557 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.061060 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.061900 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.068299 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.065064 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056887 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.059280 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.057362 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
TOIP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.926659 (rank : 2) | θ value | 6.80647e-157 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TOIP1_MOUSE
|
||||||
| NC score | 0.887408 (rank : 3) | θ value | 2.24614e-67 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921T2, Q3U7A4 | Gene names | Tor1aip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1 (Lamina-associated polypeptide 1B). | |||||
|
TOIP1_HUMAN
|
||||||
| NC score | 0.886520 (rank : 4) | θ value | 7.45998e-71 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5JTV8, Q5JTV6, Q8IZ65, Q9H8Y6, Q9HAJ1, Q9NV52, Q9Y3X5 | Gene names | TOR1AIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.155398 (rank : 5) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.152478 (rank : 6) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.143631 (rank : 7) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.129408 (rank : 8) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.128550 (rank : 9) | θ value | 0.0193708 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.120229 (rank : 10) | θ value | 0.00228821 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.117918 (rank : 11) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.095678 (rank : 12) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.094131 (rank : 13) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.093627 (rank : 14) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.093539 (rank : 15) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.089126 (rank : 16) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.087096 (rank : 17) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SH3BG_MOUSE
|
||||||
| NC score | 0.078372 (rank : 18) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.072910 (rank : 19) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.068957 (rank : 20) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.068855 (rank : 21) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.068299 (rank : 22) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.068221 (rank : 23) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.067539 (rank : 24) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.066673 (rank : 25) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.065348 (rank : 26) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.065064 (rank : 27) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.064857 (rank : 28) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.064651 (rank : 29) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.063245 (rank : 30) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.063029 (rank : 31) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.062879 (rank : 32) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.061900 (rank : 33) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.061060 (rank : 34) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.060542 (rank : 35) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.060389 (rank : 36) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.059280 (rank : 37) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.059214 (rank : 38) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.057557 (rank : 39) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.057418 (rank : 40) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.057362 (rank : 41) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.056887 (rank : 42) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.056829 (rank : 43) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.056076 (rank : 44) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.054941 (rank : 45) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.054118 (rank : 46) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.053696 (rank : 47) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.053368 (rank : 48) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.052635 (rank : 49) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.052374 (rank : 50) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.051424 (rank : 51) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.051199 (rank : 52) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.051092 (rank : 53) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.050702 (rank : 54) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.048934 (rank : 55) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.045640 (rank : 56) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.044264 (rank : 57) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.044170 (rank : 58) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.042352 (rank : 59) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
HERPU_MOUSE
|
||||||
| NC score | 0.040625 (rank : 60) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJK5 | Gene names | Herpud1 | |||
|
Domain Architecture |

|
|||||
| Description | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein. | |||||
|
S30BP_HUMAN
|
||||||
| NC score | 0.040537 (rank : 61) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHR5, Q8N1R5, Q8TDR8, Q96D27 | Gene names | SAP30BP, HCNGP, HTRG, HTRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.038431 (rank : 62) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.038160 (rank : 63) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TR10A_HUMAN
|
||||||
| NC score | 0.038056 (rank : 64) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
SH3BG_HUMAN
|
||||||
| NC score | 0.036764 (rank : 65) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55822, Q9BRB8 | Gene names | SH3BGR | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein) (21- glutamic acid-rich protein) (21-GARP). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.036236 (rank : 66) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.035978 (rank : 67) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.035919 (rank : 68) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
I28RA_HUMAN
|
||||||
| NC score | 0.035326 (rank : 69) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IU57, Q6ZML8, Q8IZI7, Q8IZI8 | Gene names | IL28RA, IFNLR1, LICR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-28 receptor alpha chain precursor (IL-28R-alpha) (IL-28RA) (Cytokine receptor family 2 member 12) (Cytokine receptor class-II member 12) (CRF2-12) (Interferon lambda receptor 1) (IFN-lambda R1) (Likely interleukin or cytokine receptor 2). | |||||
|
SHD_HUMAN
|
||||||
| NC score | 0.035146 (rank : 70) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IW2, Q96NC2 | Gene names | SHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein D. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.034444 (rank : 71) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.032428 (rank : 72) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.030966 (rank : 73) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.030287 (rank : 74) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.029729 (rank : 75) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
LMBRL_HUMAN
|
||||||
| NC score | 0.024993 (rank : 76) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UX01, Q969J4, Q96BY8, Q96HN8, Q9NT09, Q9NVE1, Q9NVU9, Q9ULP6 | Gene names | LMBR1L, KIAA1174, LIMR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LMBR1L (Lipocalin-1-interacting membrane receptor) (Lipocalin- interacting membrane receptor) (Limb region 1 protein homolog-like). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.024823 (rank : 77) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.023576 (rank : 78) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.023484 (rank : 79) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RPA1_MOUSE
|
||||||
| NC score | 0.023109 (rank : 80) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.022848 (rank : 81) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.022504 (rank : 82) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
BRMS1_HUMAN
|
||||||
| NC score | 0.022184 (rank : 83) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCU9 | Gene names | BRMS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer metastasis-suppressor 1. | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.021311 (rank : 84) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.020937 (rank : 85) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
DBR1_MOUSE
|
||||||
| NC score | 0.020811 (rank : 86) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923B1, Q8C1T9, Q8C7J7, Q99MT1 | Gene names | Dbr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lariat debranching enzyme (EC 3.1.-.-). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.020688 (rank : 87) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
LMBRL_MOUSE
|
||||||
| NC score | 0.020212 (rank : 88) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1E5, Q3TD82, Q69ZP7 | Gene names | Lmbr1l, D15Ertd735e, Kiaa1174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LMBR1L (Uteroglobin receptor). | |||||
|
ANR15_HUMAN
|
||||||
| NC score | 0.019538 (rank : 89) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
TF2AA_HUMAN
|
||||||
| NC score | 0.019477 (rank : 90) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
PSMD3_MOUSE
|
||||||
| NC score | 0.019259 (rank : 91) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14685, Q99LL7 | Gene names | Psmd3, P91a, Tstap91a | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit S3) (Proteasome subunit p58) (Transplantation antigen P91A) (Tum-P91A antigen). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.019148 (rank : 92) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.018752 (rank : 93) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.017025 (rank : 94) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
SSFA2_HUMAN
|
||||||
| NC score | 0.016967 (rank : 95) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28290, Q96E36 | Gene names | SSFA2, CS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-specific antigen 2 (Cleavage signal-1 protein) (CS-1). | |||||
|
TIM_MOUSE
|
||||||
| NC score | 0.015127 (rank : 96) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1X4, Q63ZX9, Q6P204, Q6PDL4, Q7TPV8, Q8R0Q2, Q9R268, Q9Z0E7 | Gene names | Timeless, Tim1, Timeless1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (mTim). | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.015057 (rank : 97) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.014332 (rank : 98) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.014236 (rank : 99) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.014159 (rank : 100) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
IP3KC_MOUSE
|
||||||
| NC score | 0.013697 (rank : 101) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.011561 (rank : 102) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.010968 (rank : 103) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.010461 (rank : 104) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.010419 (rank : 105) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.010343 (rank : 106) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.010161 (rank : 107) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LRCH1_MOUSE
|
||||||
| NC score | 0.008119 (rank : 108) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62046 | Gene names | Lrch1, Chdc1, Kiaa1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.007961 (rank : 109) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.007869 (rank : 110) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.007473 (rank : 111) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.006759 (rank : 112) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.006324 (rank : 113) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.005421 (rank : 114) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.004862 (rank : 115) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.003822 (rank : 116) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.002834 (rank : 117) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
GZF1_HUMAN
|
||||||
| NC score | 0.002502 (rank : 118) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H116, Q96N08, Q9BQK9, Q9H117, Q9H6W6 | Gene names | GZF1, ZBTB23, ZNF336 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF-inducible zinc finger protein 1 (Zinc finger protein 336) (Zinc finger and BTB domain-containing protein 23). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.000945 (rank : 119) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PCDG2_HUMAN
|
||||||
| NC score | 0.000779 (rank : 120) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||