Please be patient as the page loads
|
SL9A3_HUMAN
|
||||||
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
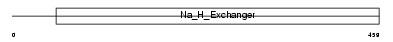
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SL9A3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
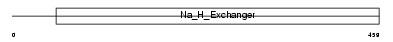
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
|
SL9A5_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981545 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
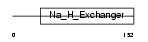
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
SL9A2_HUMAN
|
||||||
| θ value | 2.35568e-133 (rank : 3) | NC score | 0.972465 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UBY0 | Gene names | SLC9A2, NHE2 | |||
|
Domain Architecture |
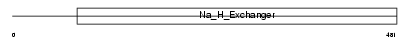
|
|||||
| Description | Sodium/hydrogen exchanger 2 (Na(+)/H(+) exchanger 2) (NHE-2) (Solute carrier family 9 member 2). | |||||
|
SL9A1_HUMAN
|
||||||
| θ value | 1.02657e-120 (rank : 4) | NC score | 0.963303 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19634 | Gene names | SLC9A1, APNH1, NHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1) (Na(+)/H(+) antiporter, amiloride- sensitive) (APNH). | |||||
|
SL9A1_MOUSE
|
||||||
| θ value | 2.28696e-120 (rank : 5) | NC score | 0.963277 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61165 | Gene names | Slc9a1, Nhe1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1). | |||||
|
SL9A9_HUMAN
|
||||||
| θ value | 9.81109e-47 (rank : 6) | NC score | 0.862061 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IVB4, Q8NAB9 | Gene names | SLC9A9, NHE9 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 9 (Na(+)/H(+) exchanger 9) (NHE-9) (Solute carrier family 9 member 9). | |||||
|
SL9A9_MOUSE
|
||||||
| θ value | 9.81109e-47 (rank : 7) | NC score | 0.862737 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BZ00, Q3U0X0 | Gene names | Slc9a9, Nhe9 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 9 (Na(+)/H(+) exchanger 9) (NHE-9) (Solute carrier family 9 member 9). | |||||
|
SL9A8_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 8) | NC score | 0.891700 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2E8, Q68CZ8, Q9BX15, Q9Y507 | Gene names | SLC9A8, KIAA0939, NHE8 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 8 (Na(+)/H(+) exchanger 8) (NHE-8) (Solute carrier family 9 member 8). | |||||
|
SL9A6_HUMAN
|
||||||
| θ value | 4.122e-45 (rank : 9) | NC score | 0.865522 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92581 | Gene names | SLC9A6, KIAA0267, NHE6 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 6 (Na(+)/H(+) exchanger 6) (NHE-6) (Solute carrier family 9 member 6). | |||||
|
SL9A8_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 10) | NC score | 0.888878 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4D1, Q3UPR4, Q5WA59, Q8BIH8, Q8BJ27 | Gene names | Slc9a8, Nhe8 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 8 (Na(+)/H(+) exchanger 8) (NHE-8) (Solute carrier family 9 member 8). | |||||
|
SL9A7_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 11) | NC score | 0.853830 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96T83, O75827, Q5JXP9 | Gene names | SLC9A7, NHE7 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 7 (Na(+)/H(+) exchanger 7) (NHE-7) (Solute carrier family 9 member 7). | |||||
|
SL9A7_MOUSE
|
||||||
| θ value | 3.05695e-40 (rank : 12) | NC score | 0.850374 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BLV3 | Gene names | Slc9a7, Nhe7 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 7 (Na(+)/H(+) exchanger 7) (NHE-7) (Solute carrier family 9 member 7). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.031969 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.018943 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.020975 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.044920 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
COFA1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.016869 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.012428 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.028132 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.024908 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.010839 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
HIAT1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.030439 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MC6, Q6P2N7, Q8N8K2, Q8NBV3, Q96NY0, Q9NT25 | Gene names | HIAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocampus abundant transcript 1 protein (Putative tetracycline transporter-like protein). | |||||
|
HIAT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.030481 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70187, Q9DBS0 | Gene names | Hiat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocampus abundant transcript 1 protein. | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.025153 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.013706 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
OR7G3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | -0.000342 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG95, Q96R99 | Gene names | OR7G3 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 7G3 (OST085). | |||||
|
ADCY7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.008339 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
LBN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.023604 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.018814 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.012318 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.013394 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.019131 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
NCOAT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.025429 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NCOAT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.025628 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.025977 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.013468 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
INSRR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | -0.000842 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTL4 | Gene names | Insrr, Irr | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor-related protein precursor (EC 2.7.10.1) (IRR) (IR- related receptor) [Contains: Insulin receptor-related protein alpha chain; Insulin receptor-related protein beta chain]. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.006251 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.009243 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.011563 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CLAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.010447 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
HEPH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.008242 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.009918 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.005296 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
O13C4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | -0.001132 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGS5, Q96R41 | Gene names | OR13C4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 13C4. | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.012349 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.008992 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.007081 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
HHCM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.017233 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05877 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocellular carcinoma protein HHCM (HHC(M)). | |||||
|
IMPG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.011750 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1W8, Q9CTP8, Q9ES62, Q9ET31 | Gene names | Impg1, Spacr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor matrix proteoglycan 1 precursor (IPM 150 proteoglycan) (Sialoprotein associated with cones and rods). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.010597 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.005466 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
PPARD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.002490 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35396, P37239 | Gene names | Ppard, Nr1c2, Pparb | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor delta (PPAR-delta) (PPAR- beta) (Nuclear hormone receptor 1) (NUC1). | |||||
|
TM7S3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.011983 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS93, Q9NUS4 | Gene names | TM7SF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane 7 superfamily member 3 precursor (Seven span transmembrane protein). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.009825 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
TSN5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.006912 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62079, O60628, O60746, Q9JLY1 | Gene names | TSPAN5, TM4SF9 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-5 (Tspan-5) (Transmembrane 4 superfamily member 9) (Tetraspan NET-4). | |||||
|
TSN5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.006912 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62080, O60628, O60746, Q9JLY1 | Gene names | Tspan5, Tm4sf9 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-5 (Tspan-5) (Transmembrane 4 superfamily member 9) (Tetraspan NET-4). | |||||
|
ANR23_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.000808 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.009114 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.008224 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.009500 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.003968 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.005433 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
SL9A3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
|
SL9A5_HUMAN
|
||||||
| NC score | 0.981545 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
SL9A2_HUMAN
|
||||||
| NC score | 0.972465 (rank : 3) | θ value | 2.35568e-133 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UBY0 | Gene names | SLC9A2, NHE2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 2 (Na(+)/H(+) exchanger 2) (NHE-2) (Solute carrier family 9 member 2). | |||||
|
SL9A1_HUMAN
|
||||||
| NC score | 0.963303 (rank : 4) | θ value | 1.02657e-120 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P19634 | Gene names | SLC9A1, APNH1, NHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1) (Na(+)/H(+) antiporter, amiloride- sensitive) (APNH). | |||||
|
SL9A1_MOUSE
|
||||||
| NC score | 0.963277 (rank : 5) | θ value | 2.28696e-120 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61165 | Gene names | Slc9a1, Nhe1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1). | |||||
|
SL9A8_HUMAN
|
||||||
| NC score | 0.891700 (rank : 6) | θ value | 1.85029e-45 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2E8, Q68CZ8, Q9BX15, Q9Y507 | Gene names | SLC9A8, KIAA0939, NHE8 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 8 (Na(+)/H(+) exchanger 8) (NHE-8) (Solute carrier family 9 member 8). | |||||
|
SL9A8_MOUSE
|
||||||
| NC score | 0.888878 (rank : 7) | θ value | 1.19932e-44 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4D1, Q3UPR4, Q5WA59, Q8BIH8, Q8BJ27 | Gene names | Slc9a8, Nhe8 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 8 (Na(+)/H(+) exchanger 8) (NHE-8) (Solute carrier family 9 member 8). | |||||
|
SL9A6_HUMAN
|
||||||
| NC score | 0.865522 (rank : 8) | θ value | 4.122e-45 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92581 | Gene names | SLC9A6, KIAA0267, NHE6 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 6 (Na(+)/H(+) exchanger 6) (NHE-6) (Solute carrier family 9 member 6). | |||||
|
SL9A9_MOUSE
|
||||||
| NC score | 0.862737 (rank : 9) | θ value | 9.81109e-47 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BZ00, Q3U0X0 | Gene names | Slc9a9, Nhe9 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 9 (Na(+)/H(+) exchanger 9) (NHE-9) (Solute carrier family 9 member 9). | |||||
|
SL9A9_HUMAN
|
||||||
| NC score | 0.862061 (rank : 10) | θ value | 9.81109e-47 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IVB4, Q8NAB9 | Gene names | SLC9A9, NHE9 | |||
|
Domain Architecture |
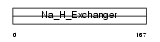
|
|||||
| Description | Sodium/hydrogen exchanger 9 (Na(+)/H(+) exchanger 9) (NHE-9) (Solute carrier family 9 member 9). | |||||
|
SL9A7_HUMAN
|
||||||
| NC score | 0.853830 (rank : 11) | θ value | 2.50074e-42 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96T83, O75827, Q5JXP9 | Gene names | SLC9A7, NHE7 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 7 (Na(+)/H(+) exchanger 7) (NHE-7) (Solute carrier family 9 member 7). | |||||
|
SL9A7_MOUSE
|
||||||
| NC score | 0.850374 (rank : 12) | θ value | 3.05695e-40 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BLV3 | Gene names | Slc9a7, Nhe7 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 7 (Na(+)/H(+) exchanger 7) (NHE-7) (Solute carrier family 9 member 7). | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.044920 (rank : 13) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.031969 (rank : 14) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
HIAT1_MOUSE
|
||||||
| NC score | 0.030481 (rank : 15) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70187, Q9DBS0 | Gene names | Hiat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocampus abundant transcript 1 protein. | |||||
|
HIAT1_HUMAN
|
||||||
| NC score | 0.030439 (rank : 16) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MC6, Q6P2N7, Q8N8K2, Q8NBV3, Q96NY0, Q9NT25 | Gene names | HIAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocampus abundant transcript 1 protein (Putative tetracycline transporter-like protein). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.028132 (rank : 17) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.025977 (rank : 18) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
NCOAT_MOUSE
|
||||||
| NC score | 0.025628 (rank : 19) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.025429 (rank : 20) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.025153 (rank : 21) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.024908 (rank : 22) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
LBN_HUMAN
|
||||||
| NC score | 0.023604 (rank : 23) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.020975 (rank : 24) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.019131 (rank : 25) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.018943 (rank : 26) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.018814 (rank : 27) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
HHCM_HUMAN
|
||||||
| NC score | 0.017233 (rank : 28) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05877 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocellular carcinoma protein HHCM (HHC(M)). | |||||
|
COFA1_MOUSE
|
||||||
| NC score | 0.016869 (rank : 29) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.013706 (rank : 30) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.013468 (rank : 31) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.013394 (rank : 32) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.012428 (rank : 33) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.012349 (rank : 34) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.012318 (rank : 35) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
TM7S3_HUMAN
|
||||||
| NC score | 0.011983 (rank : 36) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS93, Q9NUS4 | Gene names | TM7SF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane 7 superfamily member 3 precursor (Seven span transmembrane protein). | |||||
|
IMPG1_MOUSE
|
||||||
| NC score | 0.011750 (rank : 37) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1W8, Q9CTP8, Q9ES62, Q9ET31 | Gene names | Impg1, Spacr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor matrix proteoglycan 1 precursor (IPM 150 proteoglycan) (Sialoprotein associated with cones and rods). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.011563 (rank : 38) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.010839 (rank : 39) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.010597 (rank : 40) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
CLAP1_HUMAN
|
||||||
| NC score | 0.010447 (rank : 41) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.009918 (rank : 42) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.009825 (rank : 43) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.009500 (rank : 44) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.009243 (rank : 45) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.009114 (rank : 46) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.008992 (rank : 47) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ADCY7_HUMAN
|
||||||
| NC score | 0.008339 (rank : 48) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
HEPH_HUMAN
|
||||||
| NC score | 0.008242 (rank : 49) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.008224 (rank : 50) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.007081 (rank : 51) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
TSN5_HUMAN
|
||||||
| NC score | 0.006912 (rank : 52) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62079, O60628, O60746, Q9JLY1 | Gene names | TSPAN5, TM4SF9 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-5 (Tspan-5) (Transmembrane 4 superfamily member 9) (Tetraspan NET-4). | |||||
|
TSN5_MOUSE
|
||||||
| NC score | 0.006912 (rank : 53) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62080, O60628, O60746, Q9JLY1 | Gene names | Tspan5, Tm4sf9 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-5 (Tspan-5) (Transmembrane 4 superfamily member 9) (Tetraspan NET-4). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.006251 (rank : 54) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.005466 (rank : 55) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.005433 (rank : 56) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.005296 (rank : 57) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.003968 (rank : 58) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
PPARD_MOUSE
|
||||||
| NC score | 0.002490 (rank : 59) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35396, P37239 | Gene names | Ppard, Nr1c2, Pparb | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor delta (PPAR-delta) (PPAR- beta) (Nuclear hormone receptor 1) (NUC1). | |||||
|
ANR23_MOUSE
|
||||||
| NC score | 0.000808 (rank : 60) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
OR7G3_HUMAN
|
||||||
| NC score | -0.000342 (rank : 61) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG95, Q96R99 | Gene names | OR7G3 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 7G3 (OST085). | |||||
|
INSRR_MOUSE
|
||||||
| NC score | -0.000842 (rank : 62) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTL4 | Gene names | Insrr, Irr | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor-related protein precursor (EC 2.7.10.1) (IRR) (IR- related receptor) [Contains: Insulin receptor-related protein alpha chain; Insulin receptor-related protein beta chain]. | |||||
|
O13C4_HUMAN
|
||||||
| NC score | -0.001132 (rank : 63) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGS5, Q96R41 | Gene names | OR13C4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 13C4. | |||||