Please be patient as the page loads
|
NTHL1_HUMAN
|
||||||
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |
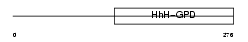
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NTHL1_HUMAN
|
||||||
| θ value | 2.18449e-163 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
NTHL1_MOUSE
|
||||||
| θ value | 6.41512e-131 (rank : 2) | NC score | 0.993801 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35980 | Gene names | Nthl1, Nth1 | |||
|
Domain Architecture |
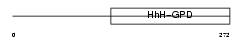
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
MUTYH_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 3) | NC score | 0.304968 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIF7, Q15830, Q9UBP2, Q9UBS7, Q9UIF4, Q9UIF5, Q9UIF6 | Gene names | MUTYH, MYH | |||
|
Domain Architecture |
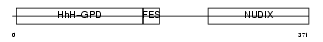
|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (hMYH). | |||||
|
MUTYH_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 4) | NC score | 0.305006 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99P21 | Gene names | Mutyh, Myh | |||
|
Domain Architecture |
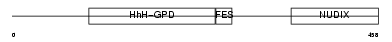
|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (mMYH). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 5) | NC score | 0.022858 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
OGG1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 6) | NC score | 0.092594 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08760, O08733, O08910, O08991, O35617, O35915, Q9QXE8 | Gene names | Ogg1 | |||
|
Domain Architecture |
|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
SDK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.011272 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.009399 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
OGG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.103806 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15527, O00390, O00670, O00705, O14876, O95488, P78554, Q9UIK0, Q9UIK1, Q9UIK2, Q9UL34, Q9Y2C0, Q9Y2C1, Q9Y6C3, Q9Y6C4 | Gene names | OGG1, MMH, MUTM, OGH1 | |||
|
Domain Architecture |
|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.031790 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
NTHL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.18449e-163 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
NTHL1_MOUSE
|
||||||
| NC score | 0.993801 (rank : 2) | θ value | 6.41512e-131 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35980 | Gene names | Nthl1, Nth1 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
MUTYH_MOUSE
|
||||||
| NC score | 0.305006 (rank : 3) | θ value | 7.1131e-05 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99P21 | Gene names | Mutyh, Myh | |||
|
Domain Architecture |

|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (mMYH). | |||||
|
MUTYH_HUMAN
|
||||||
| NC score | 0.304968 (rank : 4) | θ value | 4.1701e-05 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIF7, Q15830, Q9UBP2, Q9UBS7, Q9UIF4, Q9UIF5, Q9UIF6 | Gene names | MUTYH, MYH | |||
|
Domain Architecture |

|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (hMYH). | |||||
|
OGG1_HUMAN
|
||||||
| NC score | 0.103806 (rank : 5) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15527, O00390, O00670, O00705, O14876, O95488, P78554, Q9UIK0, Q9UIK1, Q9UIK2, Q9UL34, Q9Y2C0, Q9Y2C1, Q9Y6C3, Q9Y6C4 | Gene names | OGG1, MMH, MUTM, OGH1 | |||
|
Domain Architecture |

|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
OGG1_MOUSE
|
||||||
| NC score | 0.092594 (rank : 6) | θ value | 0.62314 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08760, O08733, O08910, O08991, O35617, O35915, Q9QXE8 | Gene names | Ogg1 | |||
|
Domain Architecture |

|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.031790 (rank : 7) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.022858 (rank : 8) | θ value | 0.163984 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
SDK2_HUMAN
|
||||||
| NC score | 0.011272 (rank : 9) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.009399 (rank : 10) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||