Please be patient as the page loads
|
OGG1_HUMAN
|
||||||
| SwissProt Accessions | O15527, O00390, O00670, O00705, O14876, O95488, P78554, Q9UIK0, Q9UIK1, Q9UIK2, Q9UL34, Q9Y2C0, Q9Y2C1, Q9Y6C3, Q9Y6C4 | Gene names | OGG1, MMH, MUTM, OGH1 | |||
|
Domain Architecture |
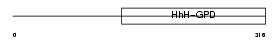
|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OGG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15527, O00390, O00670, O00705, O14876, O95488, P78554, Q9UIK0, Q9UIK1, Q9UIK2, Q9UL34, Q9Y2C0, Q9Y2C1, Q9Y6C3, Q9Y6C4 | Gene names | OGG1, MMH, MUTM, OGH1 | |||
|
Domain Architecture |

|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
OGG1_MOUSE
|
||||||
| θ value | 2.10606e-174 (rank : 2) | NC score | 0.996163 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08760, O08733, O08910, O08991, O35617, O35915, Q9QXE8 | Gene names | Ogg1 | |||
|
Domain Architecture |
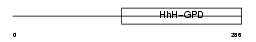
|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
NTHL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 3) | NC score | 0.121974 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35980 | Gene names | Nthl1, Nth1 | |||
|
Domain Architecture |
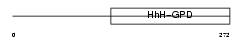
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
MUTYH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 4) | NC score | 0.078807 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P21 | Gene names | Mutyh, Myh | |||
|
Domain Architecture |
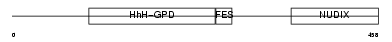
|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (mMYH). | |||||
|
NTHL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 5) | NC score | 0.103806 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |
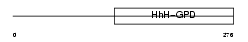
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 6) | NC score | 0.018730 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 7) | NC score | 0.019232 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CCS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 8) | NC score | 0.015441 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14618 | Gene names | CCS | |||
|
Domain Architecture |
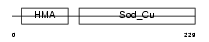
|
|||||
| Description | Copper chaperone for superoxide dismutase (Superoxide dismutase copper chaperone). | |||||
|
OGG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15527, O00390, O00670, O00705, O14876, O95488, P78554, Q9UIK0, Q9UIK1, Q9UIK2, Q9UL34, Q9Y2C0, Q9Y2C1, Q9Y6C3, Q9Y6C4 | Gene names | OGG1, MMH, MUTM, OGH1 | |||
|
Domain Architecture |

|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
OGG1_MOUSE
|
||||||
| NC score | 0.996163 (rank : 2) | θ value | 2.10606e-174 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08760, O08733, O08910, O08991, O35617, O35915, Q9QXE8 | Gene names | Ogg1 | |||
|
Domain Architecture |

|
|||||
| Description | N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]. | |||||
|
NTHL1_MOUSE
|
||||||
| NC score | 0.121974 (rank : 3) | θ value | 0.125558 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35980 | Gene names | Nthl1, Nth1 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
NTHL1_HUMAN
|
||||||
| NC score | 0.103806 (rank : 4) | θ value | 4.03905 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
MUTYH_MOUSE
|
||||||
| NC score | 0.078807 (rank : 5) | θ value | 0.163984 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P21 | Gene names | Mutyh, Myh | |||
|
Domain Architecture |

|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (mMYH). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.019232 (rank : 6) | θ value | 5.27518 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.018730 (rank : 7) | θ value | 5.27518 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CCS_HUMAN
|
||||||
| NC score | 0.015441 (rank : 8) | θ value | 8.99809 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14618 | Gene names | CCS | |||
|
Domain Architecture |

|
|||||
| Description | Copper chaperone for superoxide dismutase (Superoxide dismutase copper chaperone). | |||||