Please be patient as the page loads
|
PIGN_MOUSE
|
||||||
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PIGN_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995698 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
PIGN_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PIGG_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 3) | NC score | 0.420787 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
PIGO_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 4) | NC score | 0.316970 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 5) | NC score | 0.303752 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
ENPP6_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 6) | NC score | 0.189516 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 7) | NC score | 0.183346 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.100305 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
CTR1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.028645 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |
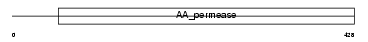
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.116715 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
GOT1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.056794 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR60 | Gene names | Golt1b | |||
|
Domain Architecture |
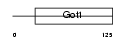
|
|||||
| Description | Vesicle transport protein GOT1B (Golgi transport 1 homolog B). | |||||
|
GOT1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.052422 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3E0, Q54A40, Q6I9W6, Q9P1R9 | Gene names | GOLT1B, GOT1A | |||
|
Domain Architecture |
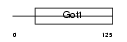
|
|||||
| Description | Vesicle transport protein GOT1B (Golgi transport 1 homolog B) (hGOT1a) (Putative NF-kappa-B-activating protein 470). | |||||
|
O11L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.001716 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGX0 | Gene names | OR11L1 | |||
|
Domain Architecture |
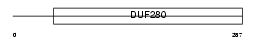
|
|||||
| Description | Olfactory receptor 11L1. | |||||
|
S45A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.032638 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2W3, Q5VY46, Q5VY49 | Gene names | SLC45A1, DNB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proton-associated sugar transporter A (PAST-A) (Solute carrier family 45 member 1) (Deleted in neuroblastoma 5 protein) (DNb-5). | |||||
|
S35B3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.022190 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922Q5 | Gene names | Slc35b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 35 member B3. | |||||
|
S45A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.031328 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIV7, Q3TZ98 | Gene names | Slc45a1, Dnb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proton-associated sugar transporter A (PAST-A) (Solute carrier family 45 member 1) (Deleted in neuroblastoma 5 protein homolog) (DNb-5 homolog). | |||||
|
UCHL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.025463 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5K5, Q5LJA6, Q5LJA7, Q8TBS4, Q96BJ9, Q9H1W5, Q9P0I3, Q9UQN2 | Gene names | UCHL5, UCH37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.013834 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DEN2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.013119 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.019556 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.018393 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.012325 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.070700 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
MOL1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.017587 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7L9L4, Q8IY23 | Gene names | MOBKL1A, MOB4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1A (Mob1 homolog 1A) (Mob1A) (Mob1B) (Protein Mob4A). | |||||
|
MOL1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.017587 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BPB0 | Gene names | Mobkl1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1A (Mob1 homolog 1A). | |||||
|
MOL1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.017586 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8S9, Q53S34, Q9H3T5, Q9HAI0, Q9NVE2 | Gene names | MOBK1B, MOB4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1B (Mob1 homolog 1B) (Mob1 alpha) (Mob1A) (Protein Mob4B). | |||||
|
MOL1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.017586 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921Y0, Q3TJA6, Q8C194, Q8C1C7 | Gene names | Mobk1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1B (Mob1 homolog 1B). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.004571 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
UCHL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.020755 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUP7, Q9CVJ4, Q9CXZ3, Q9R107 | Gene names | Uchl5, Uch37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
ENPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.061919 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.062602 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.061669 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.081363 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.080402 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.995698 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
PIGG_HUMAN
|
||||||
| NC score | 0.420787 (rank : 3) | θ value | 1.9326e-10 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
PIGO_HUMAN
|
||||||
| NC score | 0.316970 (rank : 4) | θ value | 1.69304e-06 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| NC score | 0.303752 (rank : 5) | θ value | 1.09739e-05 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
ENPP6_HUMAN
|
||||||
| NC score | 0.189516 (rank : 6) | θ value | 0.000121331 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| NC score | 0.183346 (rank : 7) | θ value | 0.00035302 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.116715 (rank : 8) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.100305 (rank : 9) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_HUMAN
|
||||||
| NC score | 0.081363 (rank : 10) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| NC score | 0.080402 (rank : 11) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.070700 (rank : 12) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.062602 (rank : 13) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.061919 (rank : 14) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.061669 (rank : 15) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
GOT1B_MOUSE
|
||||||
| NC score | 0.056794 (rank : 16) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CR60 | Gene names | Golt1b | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport protein GOT1B (Golgi transport 1 homolog B). | |||||
|
GOT1B_HUMAN
|
||||||
| NC score | 0.052422 (rank : 17) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3E0, Q54A40, Q6I9W6, Q9P1R9 | Gene names | GOLT1B, GOT1A | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport protein GOT1B (Golgi transport 1 homolog B) (hGOT1a) (Putative NF-kappa-B-activating protein 470). | |||||
|
S45A1_HUMAN
|
||||||
| NC score | 0.032638 (rank : 18) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2W3, Q5VY46, Q5VY49 | Gene names | SLC45A1, DNB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proton-associated sugar transporter A (PAST-A) (Solute carrier family 45 member 1) (Deleted in neuroblastoma 5 protein) (DNb-5). | |||||
|
S45A1_MOUSE
|
||||||
| NC score | 0.031328 (rank : 19) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BIV7, Q3TZ98 | Gene names | Slc45a1, Dnb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proton-associated sugar transporter A (PAST-A) (Solute carrier family 45 member 1) (Deleted in neuroblastoma 5 protein homolog) (DNb-5 homolog). | |||||
|
CTR1_HUMAN
|
||||||
| NC score | 0.028645 (rank : 20) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
UCHL5_HUMAN
|
||||||
| NC score | 0.025463 (rank : 21) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5K5, Q5LJA6, Q5LJA7, Q8TBS4, Q96BJ9, Q9H1W5, Q9P0I3, Q9UQN2 | Gene names | UCHL5, UCH37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
S35B3_MOUSE
|
||||||
| NC score | 0.022190 (rank : 22) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922Q5 | Gene names | Slc35b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 35 member B3. | |||||
|
UCHL5_MOUSE
|
||||||
| NC score | 0.020755 (rank : 23) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUP7, Q9CVJ4, Q9CXZ3, Q9R107 | Gene names | Uchl5, Uch37 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (EC 3.4.19.12) (UCH- L5) (Ubiquitin thioesterase L5) (Ubiquitin C-terminal hydrolase UCH37). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.019556 (rank : 24) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.018393 (rank : 25) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
MOL1A_HUMAN
|
||||||
| NC score | 0.017587 (rank : 26) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7L9L4, Q8IY23 | Gene names | MOBKL1A, MOB4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1A (Mob1 homolog 1A) (Mob1A) (Mob1B) (Protein Mob4A). | |||||
|
MOL1A_MOUSE
|
||||||
| NC score | 0.017587 (rank : 27) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BPB0 | Gene names | Mobkl1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1A (Mob1 homolog 1A). | |||||
|
MOL1B_HUMAN
|
||||||
| NC score | 0.017586 (rank : 28) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8S9, Q53S34, Q9H3T5, Q9HAI0, Q9NVE2 | Gene names | MOBK1B, MOB4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1B (Mob1 homolog 1B) (Mob1 alpha) (Mob1A) (Protein Mob4B). | |||||
|
MOL1B_MOUSE
|
||||||
| NC score | 0.017586 (rank : 29) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921Y0, Q3TJA6, Q8C194, Q8C1C7 | Gene names | Mobk1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mps one binder kinase activator-like 1B (Mob1 homolog 1B). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.013834 (rank : 30) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DEN2A_HUMAN
|
||||||
| NC score | 0.013119 (rank : 31) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.012325 (rank : 32) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.004571 (rank : 33) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
O11L1_HUMAN
|
||||||
| NC score | 0.001716 (rank : 34) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGX0 | Gene names | OR11L1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 11L1. | |||||