Please be patient as the page loads
|
PIGG_HUMAN
|
||||||
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PIGG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
PIGO_MOUSE
|
||||||
| θ value | 3.48141e-52 (rank : 2) | NC score | 0.786223 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_HUMAN
|
||||||
| θ value | 1.01294e-51 (rank : 3) | NC score | 0.786584 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGN_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 4) | NC score | 0.420787 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PIGN_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 5) | NC score | 0.407563 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
ENPP5_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 6) | NC score | 0.227968 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP6_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 7) | NC score | 0.250770 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 8) | NC score | 0.238074 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP5_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.220461 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.159566 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.138166 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
STS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.037240 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08842 | Gene names | STS | |||
|
Domain Architecture |
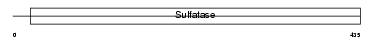
|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
ABC8A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.013931 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K442, Q6PAV3, Q8C0A9, Q8R0R4 | Gene names | Abca8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 8-A. | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.205927 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.002635 (rank : 30) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ELMO2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.016441 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHL5, Q8BHL9, Q8BQG1, Q8CBM8, Q8CC50, Q8CH98, Q91ZU2, Q9CT75 | Gene names | Elmo2, Kiaa1834 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 2 (CED-12 homolog A). | |||||
|
RPOM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.034417 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00411, O60370 | Gene names | POLRMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase, mitochondrial precursor (EC 2.7.7.6) (MtRPOL). | |||||
|
HD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.019189 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.007086 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
IGHA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.010614 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01878 | Gene names | ||||
|
Domain Architecture |
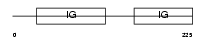
|
|||||
| Description | Ig alpha chain C region. | |||||
|
MOT4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.010693 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15427 | Gene names | SLC16A3, MCT3, MCT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 4 (MCT 4) (MCT 3) (Solute carrier family 16 member 3). | |||||
|
RPOM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.030701 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKF1, Q8BJE0 | Gene names | Polrmt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase, mitochondrial precursor (EC 2.7.7.6) (MtRPOL). | |||||
|
DEPD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.014110 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
FOXK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.005805 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |
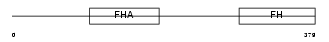
|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.005463 (rank : 29) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
TM100_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.025683 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQG9 | Gene names | Tmem100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 100. | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.007976 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
ENPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.126907 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.134067 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.131506 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
PIGG_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
PIGO_HUMAN
|
||||||
| NC score | 0.786584 (rank : 2) | θ value | 1.01294e-51 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| NC score | 0.786223 (rank : 3) | θ value | 3.48141e-52 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.420787 (rank : 4) | θ value | 1.9326e-10 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.407563 (rank : 5) | θ value | 9.59137e-10 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
ENPP6_MOUSE
|
||||||
| NC score | 0.250770 (rank : 6) | θ value | 0.000270298 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_HUMAN
|
||||||
| NC score | 0.238074 (rank : 7) | θ value | 0.000461057 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP5_HUMAN
|
||||||
| NC score | 0.227968 (rank : 8) | θ value | 9.29e-05 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| NC score | 0.220461 (rank : 9) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.205927 (rank : 10) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.159566 (rank : 11) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.138166 (rank : 12) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.134067 (rank : 13) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.131506 (rank : 14) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.126907 (rank : 15) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
STS_HUMAN
|
||||||
| NC score | 0.037240 (rank : 16) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08842 | Gene names | STS | |||
|
Domain Architecture |

|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
RPOM_HUMAN
|
||||||
| NC score | 0.034417 (rank : 17) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00411, O60370 | Gene names | POLRMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase, mitochondrial precursor (EC 2.7.7.6) (MtRPOL). | |||||
|
RPOM_MOUSE
|
||||||
| NC score | 0.030701 (rank : 18) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKF1, Q8BJE0 | Gene names | Polrmt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase, mitochondrial precursor (EC 2.7.7.6) (MtRPOL). | |||||
|
TM100_MOUSE
|
||||||
| NC score | 0.025683 (rank : 19) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQG9 | Gene names | Tmem100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 100. | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.019189 (rank : 20) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
ELMO2_MOUSE
|
||||||
| NC score | 0.016441 (rank : 21) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHL5, Q8BHL9, Q8BQG1, Q8CBM8, Q8CC50, Q8CH98, Q91ZU2, Q9CT75 | Gene names | Elmo2, Kiaa1834 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 2 (CED-12 homolog A). | |||||
|
DEPD5_HUMAN
|
||||||
| NC score | 0.014110 (rank : 22) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
ABC8A_MOUSE
|
||||||
| NC score | 0.013931 (rank : 23) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K442, Q6PAV3, Q8C0A9, Q8R0R4 | Gene names | Abca8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 8-A. | |||||
|
MOT4_HUMAN
|
||||||
| NC score | 0.010693 (rank : 24) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15427 | Gene names | SLC16A3, MCT3, MCT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 4 (MCT 4) (MCT 3) (Solute carrier family 16 member 3). | |||||
|
IGHA_MOUSE
|
||||||
| NC score | 0.010614 (rank : 25) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01878 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig alpha chain C region. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.007976 (rank : 26) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.007086 (rank : 27) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
FOXK1_MOUSE
|
||||||
| NC score | 0.005805 (rank : 28) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.005463 (rank : 29) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.002635 (rank : 30) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||