Please be patient as the page loads
|
MX2_HUMAN
|
||||||
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MX1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984718 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981954 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
MX2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.983279 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
DYN2_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 5) | NC score | 0.858573 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| θ value | 3.25095e-58 (rank : 6) | NC score | 0.858544 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 6.13105e-57 (rank : 7) | NC score | 0.851947 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DYN1_HUMAN
|
||||||
| θ value | 1.15626e-55 (rank : 8) | NC score | 0.848984 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 1.97229e-55 (rank : 9) | NC score | 0.854048 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DNM1L_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 10) | NC score | 0.877542 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
DNM1L_MOUSE
|
||||||
| θ value | 3.60269e-49 (rank : 11) | NC score | 0.876859 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 12) | NC score | 0.698452 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_MOUSE
|
||||||
| θ value | 1.5773e-20 (rank : 13) | NC score | 0.700123 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.038554 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.048509 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.039646 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
LRC49_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.036353 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YK0, Q8C3S4, Q8C7T7 | Gene names | Lrrc49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 49 (Tubulin polyglutamylase complex subunit 4) (PGs4) (p79). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.043174 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.034662 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.030548 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.029106 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.037795 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
LBN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.039855 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.033060 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.021560 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.029206 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
DP13A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.031553 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
KU86_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.028717 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27641 | Gene names | Xrcc5, G22p2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Ku autoantigen protein p86 homolog) (Ku80) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
LRC49_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.032356 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IUZ0, Q9NXM6 | Gene names | LRRC49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 49 (Tubulin polyglutamylase complex subunit 4) (PGs4). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.014253 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CTGE2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.029000 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.030899 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GCP60_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.030715 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.019488 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.029083 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.029060 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
PLCB4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.022669 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.019683 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.019560 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.024204 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
GCP60_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.028049 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.014591 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.015227 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
CA049_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.022325 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14BK3, Q4KKY9, Q9D9J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf49 homolog. | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.016451 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MFN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.080920 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IWA4, O15323, O60639, Q9BZB5, Q9NWQ2 | Gene names | MFN1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1) (Fzo homolog). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.037166 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RASF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.026122 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NHQ8, O95647, Q5SCI2, Q76KB6 | Gene names | RASSF8, C12orf2 | |||
|
Domain Architecture |
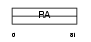
|
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1). | |||||
|
CTGE4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.024222 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
ECEL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.006412 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.026894 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.010506 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.010878 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
RCN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.030657 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |
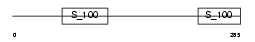
|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
S6OS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.016789 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CTN5, Q9D5F5, Q9D5Y9 | Gene names | Six6os1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SIX6OS1 (Six6 opposite strand transcript 1). | |||||
|
BCAS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.019140 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75934, Q6FGS0 | Gene names | BCAS2, DAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27). | |||||
|
BCAS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.019140 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D287, Q3TKJ5, Q91YX8, Q9D2U4, Q9DAI0 | Gene names | Bcas2, Dam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 homolog (DNA amplified in mammary carcinoma 1 protein). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.026538 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.025737 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.027958 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.025351 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CSK21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | -0.000964 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60737 | Gene names | Csnk2a1, Ckiia | |||
|
Domain Architecture |
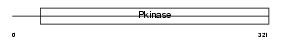
|
|||||
| Description | Casein kinase II subunit alpha (EC 2.7.11.1) (CK II). | |||||
|
CT132_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.013225 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H579, Q5JYQ6 | Gene names | C20orf132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf132. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.027336 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.000091 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.023498 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MFN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.019015 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q811U4, Q811D5, Q8CEY6, Q99M10, Q9D395 | Gene names | Mfn1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1). | |||||
|
MRP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.001725 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92887, Q14022, Q92500, Q92798, Q99663, Q9UMS2 | Gene names | ABCC2, CMOAT, CMOAT1, CMRP, MRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 1 (ATP-binding cassette sub-family C member 2) (Multidrug resistance-associated protein 2) (Canalicular multidrug resistance protein). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.020706 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.026514 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.020278 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
MX2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
MX1_HUMAN
|
||||||
| NC score | 0.984718 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| NC score | 0.983279 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| NC score | 0.981954 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
DNM1L_HUMAN
|
||||||
| NC score | 0.877542 (rank : 5) | θ value | 7.25919e-50 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
DNM1L_MOUSE
|
||||||
| NC score | 0.876859 (rank : 6) | θ value | 3.60269e-49 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
DYN2_HUMAN
|
||||||
| NC score | 0.858573 (rank : 7) | θ value | 2.48917e-58 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| NC score | 0.858544 (rank : 8) | θ value | 3.25095e-58 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.854048 (rank : 9) | θ value | 1.97229e-55 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.851947 (rank : 10) | θ value | 6.13105e-57 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.848984 (rank : 11) | θ value | 1.15626e-55 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
OPA1_MOUSE
|
||||||
| NC score | 0.700123 (rank : 12) | θ value | 1.5773e-20 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.698452 (rank : 13) | θ value | 1.5773e-20 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
MFN1_HUMAN
|
||||||
| NC score | 0.080920 (rank : 14) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IWA4, O15323, O60639, Q9BZB5, Q9NWQ2 | Gene names | MFN1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1) (Fzo homolog). | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.048509 (rank : 15) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.043174 (rank : 16) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
LBN_HUMAN
|
||||||
| NC score | 0.039855 (rank : 17) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.039646 (rank : 18) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.038554 (rank : 19) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.037795 (rank : 20) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.037166 (rank : 21) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LRC49_MOUSE
|
||||||
| NC score | 0.036353 (rank : 22) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YK0, Q8C3S4, Q8C7T7 | Gene names | Lrrc49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 49 (Tubulin polyglutamylase complex subunit 4) (PGs4) (p79). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.034662 (rank : 23) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.033060 (rank : 24) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
LRC49_HUMAN
|
||||||
| NC score | 0.032356 (rank : 25) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IUZ0, Q9NXM6 | Gene names | LRRC49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 49 (Tubulin polyglutamylase complex subunit 4) (PGs4). | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.031553 (rank : 26) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.030899 (rank : 27) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GCP60_HUMAN
|
||||||
| NC score | 0.030715 (rank : 28) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
RCN2_HUMAN
|
||||||
| NC score | 0.030657 (rank : 29) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.030548 (rank : 30) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.029206 (rank : 31) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.029106 (rank : 32) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.029083 (rank : 33) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.029060 (rank : 34) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CTGE2_HUMAN
|
||||||
| NC score | 0.029000 (rank : 35) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
KU86_MOUSE
|
||||||
| NC score | 0.028717 (rank : 36) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27641 | Gene names | Xrcc5, G22p2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Ku autoantigen protein p86 homolog) (Ku80) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
GCP60_MOUSE
|
||||||
| NC score | 0.028049 (rank : 37) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.027958 (rank : 38) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.027336 (rank : 39) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.026894 (rank : 40) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.026538 (rank : 41) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.026514 (rank : 42) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
RASF8_HUMAN
|
||||||
| NC score | 0.026122 (rank : 43) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NHQ8, O95647, Q5SCI2, Q76KB6 | Gene names | RASSF8, C12orf2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.025737 (rank : 44) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.025351 (rank : 45) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CTGE4_HUMAN
|
||||||
| NC score | 0.024222 (rank : 46) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.024204 (rank : 47) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.023498 (rank : 48) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
PLCB4_HUMAN
|
||||||
| NC score | 0.022669 (rank : 49) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
CA049_MOUSE
|
||||||
| NC score | 0.022325 (rank : 50) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14BK3, Q4KKY9, Q9D9J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf49 homolog. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.021560 (rank : 51) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.020706 (rank : 52) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.020278 (rank : 53) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.019683 (rank : 54) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.019560 (rank : 55) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.019488 (rank : 56) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
BCAS2_HUMAN
|
||||||
| NC score | 0.019140 (rank : 57) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75934, Q6FGS0 | Gene names | BCAS2, DAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27). | |||||
|
BCAS2_MOUSE
|
||||||
| NC score | 0.019140 (rank : 58) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D287, Q3TKJ5, Q91YX8, Q9D2U4, Q9DAI0 | Gene names | Bcas2, Dam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 homolog (DNA amplified in mammary carcinoma 1 protein). | |||||
|
MFN1_MOUSE
|
||||||
| NC score | 0.019015 (rank : 59) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q811U4, Q811D5, Q8CEY6, Q99M10, Q9D395 | Gene names | Mfn1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1). | |||||
|
S6OS1_MOUSE
|
||||||
| NC score | 0.016789 (rank : 60) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CTN5, Q9D5F5, Q9D5Y9 | Gene names | Six6os1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SIX6OS1 (Six6 opposite strand transcript 1). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.016451 (rank : 61) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.015227 (rank : 62) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.014591 (rank : 63) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.014253 (rank : 64) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CT132_HUMAN
|
||||||
| NC score | 0.013225 (rank : 65) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H579, Q5JYQ6 | Gene names | C20orf132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf132. | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.010878 (rank : 66) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.010506 (rank : 67) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ECEL1_MOUSE
|
||||||
| NC score | 0.006412 (rank : 68) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
MRP2_HUMAN
|
||||||
| NC score | 0.001725 (rank : 69) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92887, Q14022, Q92500, Q92798, Q99663, Q9UMS2 | Gene names | ABCC2, CMOAT, CMOAT1, CMRP, MRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Canalicular multispecific organic anion transporter 1 (ATP-binding cassette sub-family C member 2) (Multidrug resistance-associated protein 2) (Canalicular multidrug resistance protein). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.000091 (rank : 70) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
CSK21_MOUSE
|
||||||
| NC score | -0.000964 (rank : 71) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60737 | Gene names | Csnk2a1, Ckiia | |||
|
Domain Architecture |

|
|||||
| Description | Casein kinase II subunit alpha (EC 2.7.11.1) (CK II). | |||||