Please be patient as the page loads
|
MTA1_HUMAN
|
||||||
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |
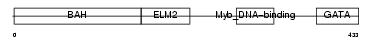
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999148 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |
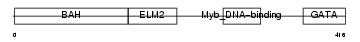
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.990179 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.989374 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992455 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |
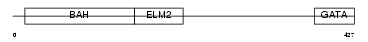
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.992039 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
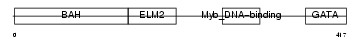
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 3.73686e-38 (rank : 7) | NC score | 0.583820 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 8) | NC score | 0.589275 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 9) | NC score | 0.494881 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 10) | NC score | 0.500051 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 11) | NC score | 0.439946 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 12) | NC score | 0.470958 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 13) | NC score | 0.494241 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 14) | NC score | 0.445280 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 15) | NC score | 0.444162 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 16) | NC score | 0.483346 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 17) | NC score | 0.280524 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 18) | NC score | 0.280181 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 19) | NC score | 0.338012 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.125922 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.011865 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
RL32_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.046011 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62910, P02433 | Gene names | RPL32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L32. | |||||
|
RL32_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.046011 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62911, P02433, Q3UFJ7 | Gene names | Rpl32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L32. | |||||
|
RTP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.026994 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DX8, Q9H4F3 | Gene names | RTP4, IFRG28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 4 (28 kDa interferon-responsive protein). | |||||
|
CJ108_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.055357 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
LRRC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.005229 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYS8, Q96LT5 | Gene names | LRRC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 2. | |||||
|
PRGC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.012366 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.008856 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PITX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.004660 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |
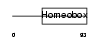
|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
STK16_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.000736 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88697, Q9JMJ0, Q9JMJ1, Q9QX00 | Gene names | Stk16, Edpk, Krct, Mpsk1, Pkl12, Tsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 16 (EC 2.7.11.1) (Protein kinase PKL12) (Myristoylated and palmitoylated serine/threonine-protein kinase) (MPSK) (TGF-beta-stimulated factor 1) (TSF-1) (Protein kinase Krct). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.006610 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.010984 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.001458 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.051328 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.000764 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
INHBE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.003266 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58166 | Gene names | INHBE | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin beta E chain precursor (Activin beta-E chain). | |||||
|
IQEC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.003930 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0S2, Q3TZC3, Q5DU15 | Gene names | Iqsec1, Kiaa0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1. | |||||
|
LRTM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.000709 (rank : 45) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XG9, Q8C030, Q8JZS8 | Gene names | Lrrtm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 4 precursor. | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | -0.001875 (rank : 46) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
S40A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.017552 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHI9, Q3UHZ9, Q8BME5, Q8BUM5, Q9JIM9, Q9JKP4 | Gene names | Slc40a1, Fpn1, Ireg1, Slc11a3, Slc39a1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 40 member 1 (Ferroportin-1) (Iron-regulated transporter 1) (Metal transporter protein 1) (MTP1). | |||||
|
VP13A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.013255 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RL7, Q5JSX9, Q5JSY0, Q5VYR5, Q702P4, Q709D0, Q86YF8, Q96S61, Q9H995, Q9Y2J1 | Gene names | VPS13A, CHAC, KIAA0986 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 13A (Chorein) (Chorea- acanthocytosis protein). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.012467 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.132166 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.079083 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.079230 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.082272 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MTA1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| NC score | 0.999148 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA3_HUMAN
|
||||||
| NC score | 0.992455 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.992039 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.990179 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.989374 (rank : 6) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.589275 (rank : 7) | θ value | 1.08726e-37 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.583820 (rank : 8) | θ value | 3.73686e-38 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.500051 (rank : 9) | θ value | 6.64225e-11 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.494881 (rank : 10) | θ value | 6.64225e-11 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.494241 (rank : 11) | θ value | 1.53129e-07 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.483346 (rank : 12) | θ value | 5.81887e-07 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.470958 (rank : 13) | θ value | 4.0297e-08 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.445280 (rank : 14) | θ value | 4.45536e-07 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.444162 (rank : 15) | θ value | 4.45536e-07 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.439946 (rank : 16) | θ value | 4.0297e-08 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.338012 (rank : 17) | θ value | 0.000270298 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.280524 (rank : 18) | θ value | 1.09739e-05 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.280181 (rank : 19) | θ value | 5.44631e-05 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.132166 (rank : 20) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.125922 (rank : 21) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.082272 (rank : 22) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.079230 (rank : 23) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.079083 (rank : 24) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
CJ108_HUMAN
|
||||||
| NC score | 0.055357 (rank : 25) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.051328 (rank : 26) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
RL32_HUMAN
|
||||||
| NC score | 0.046011 (rank : 27) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62910, P02433 | Gene names | RPL32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L32. | |||||
|
RL32_MOUSE
|
||||||
| NC score | 0.046011 (rank : 28) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62911, P02433, Q3UFJ7 | Gene names | Rpl32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L32. | |||||
|
RTP4_HUMAN
|
||||||
| NC score | 0.026994 (rank : 29) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DX8, Q9H4F3 | Gene names | RTP4, IFRG28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 4 (28 kDa interferon-responsive protein). | |||||
|
S40A1_MOUSE
|
||||||
| NC score | 0.017552 (rank : 30) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHI9, Q3UHZ9, Q8BME5, Q8BUM5, Q9JIM9, Q9JKP4 | Gene names | Slc40a1, Fpn1, Ireg1, Slc11a3, Slc39a1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 40 member 1 (Ferroportin-1) (Iron-regulated transporter 1) (Metal transporter protein 1) (MTP1). | |||||
|
VP13A_HUMAN
|
||||||
| NC score | 0.013255 (rank : 31) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RL7, Q5JSX9, Q5JSY0, Q5VYR5, Q702P4, Q709D0, Q86YF8, Q96S61, Q9H995, Q9Y2J1 | Gene names | VPS13A, CHAC, KIAA0986 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 13A (Chorein) (Chorea- acanthocytosis protein). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.012467 (rank : 32) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
PRGC1_HUMAN
|
||||||
| NC score | 0.012366 (rank : 33) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.011865 (rank : 34) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.010984 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.008856 (rank : 36) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.006610 (rank : 37) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
LRRC2_HUMAN
|
||||||
| NC score | 0.005229 (rank : 38) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYS8, Q96LT5 | Gene names | LRRC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 2. | |||||
|
PITX2_MOUSE
|
||||||
| NC score | 0.004660 (rank : 39) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97474, O08646, O70336, P97933, Q9JLA0, Q9QXB8, Q9R1V9, Q9Z141 | Gene names | Pitx2, Arp1, Brx1, Otlx2, Ptx2, Rgs | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary homeobox 2 (Orthodenticle-like homeobox 2) (Solurshin) (ALL1-responsive protein ARP1) (BRX1 homeoprotein) (Paired-like homeodomain transcription factor Munc 30). | |||||
|
IQEC1_MOUSE
|
||||||
| NC score | 0.003930 (rank : 40) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0S2, Q3TZC3, Q5DU15 | Gene names | Iqsec1, Kiaa0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1. | |||||
|
INHBE_HUMAN
|
||||||
| NC score | 0.003266 (rank : 41) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58166 | Gene names | INHBE | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin beta E chain precursor (Activin beta-E chain). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.001458 (rank : 42) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.000764 (rank : 43) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
STK16_MOUSE
|
||||||
| NC score | 0.000736 (rank : 44) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88697, Q9JMJ0, Q9JMJ1, Q9QX00 | Gene names | Stk16, Edpk, Krct, Mpsk1, Pkl12, Tsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 16 (EC 2.7.11.1) (Protein kinase PKL12) (Myristoylated and palmitoylated serine/threonine-protein kinase) (MPSK) (TGF-beta-stimulated factor 1) (TSF-1) (Protein kinase Krct). | |||||
|
LRTM4_MOUSE
|
||||||
| NC score | 0.000709 (rank : 45) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XG9, Q8C030, Q8JZS8 | Gene names | Lrrtm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 4 precursor. | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | -0.001875 (rank : 46) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||