Please be patient as the page loads
|
THAP1_MOUSE
|
||||||
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |
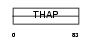
|
|||||
| Description | THAP domain-containing protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
THAP1_MOUSE
|
||||||
| θ value | 1.14028e-103 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP1_HUMAN
|
||||||
| θ value | 4.19693e-98 (rank : 2) | NC score | 0.992186 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NVV9, Q53FQ1, Q6IA99 | Gene names | THAP1 | |||
|
Domain Architecture |
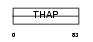
|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP2_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 3) | NC score | 0.827669 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |
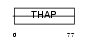
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP2_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 4) | NC score | 0.819212 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0W7 | Gene names | THAP2 | |||
|
Domain Architecture |
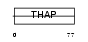
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP3_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 5) | NC score | 0.819464 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP3_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 6) | NC score | 0.821492 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJ25, Q8BII6 | Gene names | Thap3 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 7) | NC score | 0.676378 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 8) | NC score | 0.668262 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP6_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 9) | NC score | 0.675855 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
THAP8_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 10) | NC score | 0.685374 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.492975 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.485649 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
P52K_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.276383 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
P52K_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.282179 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.048003 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.041390 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.057327 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
THA11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.184860 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK4, O94795 | Gene names | THAP11 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
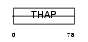
THA11_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.167809 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJD0 | Gene names | Thap11 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.040052 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.039635 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.040546 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.040203 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.053400 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.037850 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.036746 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.038738 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CF182_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.052181 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYX8 | Gene names | C6orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182. | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.018808 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.034463 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.035417 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.023215 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.038633 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.025181 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.033019 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.041494 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.041145 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.030625 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.028647 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.029759 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
TRI22_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.005315 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYM9, Q15521 | Gene names | TRIM22, RNF94, STAF50 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 22 (RING finger protein 94) (50 kDa-stimulated trans-acting factor) (Staf-50). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.043347 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CF182_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.044312 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
CCDC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.016414 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQI4 | Gene names | CCDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 3 precursor. | |||||
|
CEP70_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.039579 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.034261 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
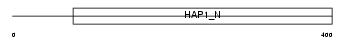
HAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.037582 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |
|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.034106 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.036266 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
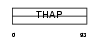
THA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.193799 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
THAP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.14028e-103 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP1_HUMAN
|
||||||
| NC score | 0.992186 (rank : 2) | θ value | 4.19693e-98 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NVV9, Q53FQ1, Q6IA99 | Gene names | THAP1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP2_MOUSE
|
||||||
| NC score | 0.827669 (rank : 3) | θ value | 2.43343e-21 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP3_MOUSE
|
||||||
| NC score | 0.821492 (rank : 4) | θ value | 2.97466e-19 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJ25, Q8BII6 | Gene names | Thap3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP3_HUMAN
|
||||||
| NC score | 0.819464 (rank : 5) | θ value | 1.33526e-19 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP2_HUMAN
|
||||||
| NC score | 0.819212 (rank : 6) | θ value | 1.5773e-20 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0W7 | Gene names | THAP2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP8_HUMAN
|
||||||
| NC score | 0.685374 (rank : 7) | θ value | 6.21693e-09 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.676378 (rank : 8) | θ value | 4.44505e-15 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP6_HUMAN
|
||||||
| NC score | 0.675855 (rank : 9) | θ value | 7.34386e-10 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.668262 (rank : 10) | θ value | 4.44505e-15 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.492975 (rank : 11) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_MOUSE
|
||||||
| NC score | 0.485649 (rank : 12) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
P52K_MOUSE
|
||||||
| NC score | 0.282179 (rank : 13) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.276383 (rank : 14) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
THA10_HUMAN
|
||||||
| NC score | 0.193799 (rank : 15) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
THA11_HUMAN
|
||||||
| NC score | 0.184860 (rank : 16) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK4, O94795 | Gene names | THAP11 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THA11_MOUSE
|
||||||
| NC score | 0.167809 (rank : 17) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJD0 | Gene names | Thap11 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.057327 (rank : 18) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.053400 (rank : 19) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CF182_HUMAN
|
||||||
| NC score | 0.052181 (rank : 20) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYX8 | Gene names | C6orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.048003 (rank : 21) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CF182_MOUSE
|
||||||
| NC score | 0.044312 (rank : 22) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.043347 (rank : 23) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.041494 (rank : 24) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.041390 (rank : 25) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.041145 (rank : 26) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.040546 (rank : 27) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.040203 (rank : 28) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.040052 (rank : 29) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.039635 (rank : 30) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
CEP70_MOUSE
|
||||||
| NC score | 0.039579 (rank : 31) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.038738 (rank : 32) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.038633 (rank : 33) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.037850 (rank : 34) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HAP1_MOUSE
|
||||||
| NC score | 0.037582 (rank : 35) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.036746 (rank : 36) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.036266 (rank : 37) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.035417 (rank : 38) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.034463 (rank : 39) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.034261 (rank : 40) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.034106 (rank : 41) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.033019 (rank : 42) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.030625 (rank : 43) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.029759 (rank : 44) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.028647 (rank : 45) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.025181 (rank : 46) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.023215 (rank : 47) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.018808 (rank : 48) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
CCDC3_HUMAN
|
||||||
| NC score | 0.016414 (rank : 49) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQI4 | Gene names | CCDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 3 precursor. | |||||
|
TRI22_HUMAN
|
||||||
| NC score | 0.005315 (rank : 50) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYM9, Q15521 | Gene names | TRIM22, RNF94, STAF50 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 22 (RING finger protein 94) (50 kDa-stimulated trans-acting factor) (Staf-50). | |||||