Please be patient as the page loads
|
ITPR2_HUMAN
|
||||||
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITPR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993064 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992587 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998677 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.990631 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.991231 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 7) | NC score | 0.534861 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 8) | NC score | 0.541309 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 9) | NC score | 0.530373 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.028142 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.018489 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.037665 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.029617 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.027534 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.017736 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CP2AC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.010537 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56593 | Gene names | Cyp2a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2A12 (EC 1.14.14.1) (CYPIIA12) (Steroid hormones 7- alpha-hydroxylase) (Testosterone 7-alpha-hydroxylase). | |||||
|
LRBA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.019212 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.018430 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.028305 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
P2RY1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.005235 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49650 | Gene names | P2ry1 | |||
|
Domain Architecture |
|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.032163 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
GA45B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.028879 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22339, Q3U342, Q9R106 | Gene names | Gadd45b, Myd118 | |||
|
Domain Architecture |

|
|||||
| Description | Growth arrest and DNA-damage-inducible protein GADD45 beta (Negative growth-regulatory protein MyD118) (Myeloid differentiation primary response protein MyD118). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.010598 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.013574 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CEP72_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.026049 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D3R3, Q8BM53, Q9D0B7 | Gene names | Cep72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.008962 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
P2RY1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.002503 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47900 | Gene names | P2RY1 | |||
|
Domain Architecture |
|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.020944 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CP2A6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.007355 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11509, P00190, P10890, Q16803, Q4VAT9, Q4VAU0, Q4VAU1, Q9H1Z7, Q9UK48 | Gene names | CYP2A6 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2A6 (EC 1.14.14.1) (CYPIIA6) (Coumarin 7-hydroxylase) (P450 IIA3) (CYP2A3) (P450(I)). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.008530 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.007932 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
LRRC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.007316 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
S26A6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.012687 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXS9, Q96Q90 | Gene names | SLC26A6 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 26 member 6 (Pendrin-like protein 1) (Pendrin L1). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.008621 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.023196 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MIS12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.026654 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H081, Q96N24 | Gene names | MIS12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MIS12 homolog. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.014689 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
UBR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.008481 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWV8, O15057, Q4VXK2, Q5TFH6, Q6P2I2, Q6ZUD0 | Gene names | UBR2, C6orf133, KIAA0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
ANGP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.007243 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.006842 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08538 | Gene names | Angpt1, Agpt | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ASB17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.026708 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHP9, Q9CUF9 | Gene names | Asb17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 17 (ASB-17). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.010045 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.011696 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.010077 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
ELMD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.009709 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N336, Q9NPW3 | Gene names | ELMOD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELMO domain-containing protein 1. | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.010865 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.012083 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.019754 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.016885 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
VDP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.020722 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.007479 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.011744 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.017368 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.030734 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.018982 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.009152 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
LRRC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.005235 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
PI3R6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.013506 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5UE93, Q658R3 | Gene names | PIK3R6, C17orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 6 (Phosphoinositide 3- kinase gamma adapter protein of 87 kDa) (p87 PI3K adapter protein) (p87PIKAP). | |||||
|
SLIT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.000140 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R1B9, Q9Z166 | Gene names | Slit2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 2 protein precursor (Slit-2) [Contains: Slit homolog 2 protein N-product; Slit homolog 2 protein C-product]. | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.015127 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.004320 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
WDTC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.005910 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
CKAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.016413 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
EMIL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.006725 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.000118 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.011493 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
KAD7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.020293 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.007850 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
O10W1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | -0.001739 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGF6, Q6UXQ2 | Gene names | OR10W1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 10W1 (Olfactory receptor OR11-236). | |||||
|
OL139_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | -0.000524 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60891, Q8VGR1, Q9EPY8 | Gene names | Olfr139, Mor255-2, Olfr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 139 (Olfactory receptor 1) (Olfactory receptor 255- 2) (Odorant receptor M5). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.005899 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SYF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.009381 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D198 | Gene names | Syf2, Cbpin, Gcipip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor SYF2 (CCNDBP1-interactor) (p29) (mp29). | |||||
|
TERF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.006827 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
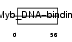
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
WDTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.005290 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
ITPR2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.998677 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.993064 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.992587 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.991231 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_HUMAN
|
||||||
| NC score | 0.990631 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.541309 (rank : 7) | θ value | 1.33217e-27 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.534861 (rank : 8) | θ value | 7.80994e-28 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.530373 (rank : 9) | θ value | 1.12775e-26 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.037665 (rank : 10) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.032163 (rank : 11) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.030734 (rank : 12) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.029617 (rank : 13) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
GA45B_MOUSE
|
||||||
| NC score | 0.028879 (rank : 14) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22339, Q3U342, Q9R106 | Gene names | Gadd45b, Myd118 | |||
|
Domain Architecture |

|
|||||
| Description | Growth arrest and DNA-damage-inducible protein GADD45 beta (Negative growth-regulatory protein MyD118) (Myeloid differentiation primary response protein MyD118). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.028305 (rank : 15) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.028142 (rank : 16) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.027534 (rank : 17) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
ASB17_MOUSE
|
||||||
| NC score | 0.026708 (rank : 18) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHP9, Q9CUF9 | Gene names | Asb17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 17 (ASB-17). | |||||
|
MIS12_HUMAN
|
||||||
| NC score | 0.026654 (rank : 19) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H081, Q96N24 | Gene names | MIS12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MIS12 homolog. | |||||
|
CEP72_MOUSE
|
||||||
| NC score | 0.026049 (rank : 20) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D3R3, Q8BM53, Q9D0B7 | Gene names | Cep72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.023196 (rank : 21) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.020944 (rank : 22) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.020722 (rank : 23) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
KAD7_HUMAN
|
||||||
| NC score | 0.020293 (rank : 24) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.019754 (rank : 25) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.019212 (rank : 26) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.018982 (rank : 27) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.018489 (rank : 28) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.018430 (rank : 29) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.017736 (rank : 30) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.017368 (rank : 31) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.016885 (rank : 32) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
CKAP2_MOUSE
|
||||||
| NC score | 0.016413 (rank : 33) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.015127 (rank : 34) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.014689 (rank : 35) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.013574 (rank : 36) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
PI3R6_HUMAN
|
||||||
| NC score | 0.013506 (rank : 37) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5UE93, Q658R3 | Gene names | PIK3R6, C17orf38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 6 (Phosphoinositide 3- kinase gamma adapter protein of 87 kDa) (p87 PI3K adapter protein) (p87PIKAP). | |||||
|
S26A6_HUMAN
|
||||||
| NC score | 0.012687 (rank : 38) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXS9, Q96Q90 | Gene names | SLC26A6 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 26 member 6 (Pendrin-like protein 1) (Pendrin L1). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.012083 (rank : 39) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.011744 (rank : 40) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.011696 (rank : 41) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.011493 (rank : 42) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.010865 (rank : 43) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.010598 (rank : 44) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
CP2AC_MOUSE
|
||||||
| NC score | 0.010537 (rank : 45) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56593 | Gene names | Cyp2a12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome P450 2A12 (EC 1.14.14.1) (CYPIIA12) (Steroid hormones 7- alpha-hydroxylase) (Testosterone 7-alpha-hydroxylase). | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.010077 (rank : 46) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
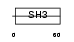
CASL_HUMAN
|
||||||
| NC score | 0.010045 (rank : 47) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
ELMD1_HUMAN
|
||||||
| NC score | 0.009709 (rank : 48) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N336, Q9NPW3 | Gene names | ELMOD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELMO domain-containing protein 1. | |||||
|
SYF2_MOUSE
|
||||||
| NC score | 0.009381 (rank : 49) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D198 | Gene names | Syf2, Cbpin, Gcipip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor SYF2 (CCNDBP1-interactor) (p29) (mp29). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.009152 (rank : 50) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.008962 (rank : 51) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.008621 (rank : 52) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.008530 (rank : 53) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
UBR2_HUMAN
|
||||||
| NC score | 0.008481 (rank : 54) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWV8, O15057, Q4VXK2, Q5TFH6, Q6P2I2, Q6ZUD0 | Gene names | UBR2, C6orf133, KIAA0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.007932 (rank : 55) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.007850 (rank : 56) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.007479 (rank : 57) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
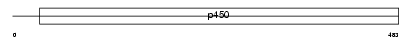
CP2A6_HUMAN
|
||||||
| NC score | 0.007355 (rank : 58) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11509, P00190, P10890, Q16803, Q4VAT9, Q4VAU0, Q4VAU1, Q9H1Z7, Q9UK48 | Gene names | CYP2A6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2A6 (EC 1.14.14.1) (CYPIIA6) (Coumarin 7-hydroxylase) (P450 IIA3) (CYP2A3) (P450(I)). | |||||
|
LRRC1_MOUSE
|
||||||
| NC score | 0.007316 (rank : 59) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
ANGP1_HUMAN
|
||||||
| NC score | 0.007243 (rank : 60) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15389 | Gene names | ANGPT1, KIAA0003 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
ANGP1_MOUSE
|
||||||
| NC score | 0.006842 (rank : 61) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08538 | Gene names | Angpt1, Agpt | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 precursor (ANG-1). | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.006827 (rank : 62) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
EMIL1_HUMAN
|
||||||
| NC score | 0.006725 (rank : 63) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
WDTC1_HUMAN
|
||||||
| NC score | 0.005910 (rank : 64) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5D0, Q9NV87, Q9UPW4 | Gene names | WDTC1, KIAA1037 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.005899 (rank : 65) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
WDTC1_MOUSE
|
||||||
| NC score | 0.005290 (rank : 66) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80ZK9 | Gene names | Wdtc1 | |||
|
Domain Architecture |

|
|||||
| Description | WD and tetratricopeptide repeats protein 1. | |||||
|
LRRC1_HUMAN
|
||||||
| NC score | 0.005235 (rank : 67) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
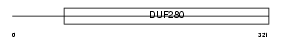
P2RY1_MOUSE
|
||||||
| NC score | 0.005235 (rank : 68) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49650 | Gene names | P2ry1 | |||
|
Domain Architecture |

|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.004320 (rank : 69) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
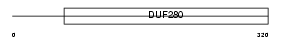
P2RY1_HUMAN
|
||||||
| NC score | 0.002503 (rank : 70) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47900 | Gene names | P2RY1 | |||
|
Domain Architecture |

|
|||||
| Description | P2Y purinoceptor 1 (ATP receptor) (P2Y1) (Purinergic receptor). | |||||
|
SLIT2_MOUSE
|
||||||
| NC score | 0.000140 (rank : 71) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R1B9, Q9Z166 | Gene names | Slit2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 2 protein precursor (Slit-2) [Contains: Slit homolog 2 protein N-product; Slit homolog 2 protein C-product]. | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.000118 (rank : 72) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
OL139_MOUSE
|
||||||
| NC score | -0.000524 (rank : 73) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60891, Q8VGR1, Q9EPY8 | Gene names | Olfr139, Mor255-2, Olfr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 139 (Olfactory receptor 1) (Olfactory receptor 255- 2) (Odorant receptor M5). | |||||
|
O10W1_HUMAN
|
||||||
| NC score | -0.001739 (rank : 74) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGF6, Q6UXQ2 | Gene names | OR10W1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 10W1 (Olfactory receptor OR11-236). | |||||