Please be patient as the page loads
|
S12A7_MOUSE
|
||||||
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S12A4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995361 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995523 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996431 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996350 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994345 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.994251 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A7_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.998290 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A2_HUMAN
|
||||||
| θ value | 3.84025e-59 (rank : 9) | NC score | 0.815241 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A3_MOUSE
|
||||||
| θ value | 3.84025e-59 (rank : 10) | NC score | 0.839373 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| θ value | 6.55044e-59 (rank : 11) | NC score | 0.816293 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| θ value | 5.54529e-58 (rank : 12) | NC score | 0.822137 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 13) | NC score | 0.807788 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| θ value | 4.11246e-53 (rank : 14) | NC score | 0.807690 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
LAT2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 15) | NC score | 0.133207 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
LAT2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 16) | NC score | 0.132749 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 17) | NC score | 0.221858 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.157260 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |
|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.152299 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.115560 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
CTR2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.197552 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |
|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.186933 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.170440 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
AAA1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.102364 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |
|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
BAT1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.139699 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
AAA1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.109053 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
LIN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.018014 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
BAT1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.121443 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.007585 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
OL998_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | -0.000066 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 648 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VF76 | Gene names | Olfr998, Mor175-5 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 998 (Olfactory receptor 175-5). | |||||
|
OR8U1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | -0.000058 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NH10 | Gene names | OR8U1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 8U1. | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.000866 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
GPR84_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.009460 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQS5 | Gene names | GPR84, EX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 84 (Inflammation-related G-protein coupled receptor EX33). | |||||
|
O1044_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | -0.000150 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VGR9 | Gene names | Olfr1044, Mor185-4 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 1044 (Olfactory receptor 185-4). | |||||
|
OR8B8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | -0.000149 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15620, Q96RC8 | Gene names | OR8B8 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 8B8 (Olfactory receptor TPCR85) (Olfactory-like receptor JCG8). | |||||
|
LAT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.076359 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z127, Q9JMI4 | Gene names | Slc7a5, Lat1 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC). | |||||
|
NBEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.011657 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.011549 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
RHBD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.010318 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |
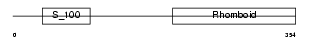
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
GP149_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.009299 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UVY1, Q3TMB1, Q80T52, Q8BXA3, Q8BZC0 | Gene names | Gpr149, Pgr10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
IRK8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.002842 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15842, O00657 | Gene names | KCNJ8 | |||
|
Domain Architecture |
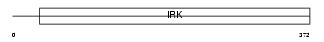
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
|
LAT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.080029 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01650, Q9UBN8, Q9UP15, Q9UQC0 | Gene names | SLC7A5, CD98LC, LAT1, MPE16 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC) (CD98 light chain) (Integral membrane protein E16) (hLAT1). | |||||
|
O1052_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.001679 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VGR8 | Gene names | Olfr1052, Mor172-1 | |||
|
Domain Architecture |
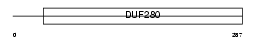
|
|||||
| Description | Olfactory receptor 1052 (Olfactory receptor 172-1). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.017030 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
F13B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.004929 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | -0.000310 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
KLF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | -0.002131 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95600, Q59GV5, Q5HYQ5, Q5JXP7, Q6MZJ7, Q9UGC4 | Gene names | KLF8, BKLF3, ZNF741 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 8 (Basic krueppel-like factor 3) (Zinc finger protein 741). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.016855 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ZFP95_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | -0.002844 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1D8 | Gene names | Zfp95 | |||
|
Domain Architecture |
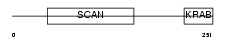
|
|||||
| Description | Zinc finger protein 95 (Zfp-95). | |||||
|
5HT1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | -0.001053 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28334 | Gene names | Htr1b, 5ht1b | |||
|
Domain Architecture |
|
|||||
| Description | 5-hydroxytryptamine 1B receptor (5-HT-1B) (Serotonin receptor 1B) (5- HT1B). | |||||
|
CT098_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.011676 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZP1, Q6FII5, Q9NUD3 | Gene names | C20orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf98. | |||||
|
EDG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.005718 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08530, Q9R235 | Gene names | Edg1, Lpb1 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1) (Lysophospholipid receptor B1). | |||||
|
F13B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.004348 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
G137B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.049825 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
RHBD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.008354 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
YLA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.077864 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UM01, O95984, Q9P2V5 | Gene names | SLC7A7 | |||
|
Domain Architecture |
|
|||||
| Description | Y+L amino acid transporter 1 (y(+)L-type amino acid transporter 1) (y+LAT-1) (Y+LAT1) (Monocyte amino acid permease 2) (MOP-2). | |||||
|
XCT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.071797 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPY5 | Gene names | SLC7A11 | |||
|
Domain Architecture |
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT) (Calcium channel blocker resistance protein CCBR1). | |||||
|
XCT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.077878 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_HUMAN
|
||||||
| NC score | 0.998290 (rank : 2) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A5_HUMAN
|
||||||
| NC score | 0.996431 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.996350 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A4_MOUSE
|
||||||
| NC score | 0.995523 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A4_HUMAN
|
||||||
| NC score | 0.995361 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.994345 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| NC score | 0.994251 (rank : 8) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A3_MOUSE
|
||||||
| NC score | 0.839373 (rank : 9) | θ value | 3.84025e-59 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| NC score | 0.822137 (rank : 10) | θ value | 5.54529e-58 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| NC score | 0.816293 (rank : 11) | θ value | 6.55044e-59 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| NC score | 0.815241 (rank : 12) | θ value | 3.84025e-59 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| NC score | 0.807788 (rank : 13) | θ value | 1.08223e-53 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| NC score | 0.807690 (rank : 14) | θ value | 4.11246e-53 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.221858 (rank : 15) | θ value | 0.00102713 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR2_HUMAN
|
||||||
| NC score | 0.197552 (rank : 16) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_MOUSE
|
||||||
| NC score | 0.186933 (rank : 17) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR1_HUMAN
|
||||||
| NC score | 0.170440 (rank : 18) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR3_HUMAN
|
||||||
| NC score | 0.157260 (rank : 19) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.152299 (rank : 20) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
BAT1_MOUSE
|
||||||
| NC score | 0.139699 (rank : 21) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
LAT2_HUMAN
|
||||||
| NC score | 0.133207 (rank : 22) | θ value | 0.00020696 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
LAT2_MOUSE
|
||||||
| NC score | 0.132749 (rank : 23) | θ value | 0.000270298 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
BAT1_HUMAN
|
||||||
| NC score | 0.121443 (rank : 24) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR4_HUMAN
|
||||||
| NC score | 0.115560 (rank : 25) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
AAA1_MOUSE
|
||||||
| NC score | 0.109053 (rank : 26) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
AAA1_HUMAN
|
||||||
| NC score | 0.102364 (rank : 27) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |

|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
LAT1_HUMAN
|
||||||
| NC score | 0.080029 (rank : 28) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01650, Q9UBN8, Q9UP15, Q9UQC0 | Gene names | SLC7A5, CD98LC, LAT1, MPE16 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC) (CD98 light chain) (Integral membrane protein E16) (hLAT1). | |||||
|
XCT_MOUSE
|
||||||
| NC score | 0.077878 (rank : 29) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
YLA1_HUMAN
|
||||||
| NC score | 0.077864 (rank : 30) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UM01, O95984, Q9P2V5 | Gene names | SLC7A7 | |||
|
Domain Architecture |

|
|||||
| Description | Y+L amino acid transporter 1 (y(+)L-type amino acid transporter 1) (y+LAT-1) (Y+LAT1) (Monocyte amino acid permease 2) (MOP-2). | |||||
|
LAT1_MOUSE
|
||||||
| NC score | 0.076359 (rank : 31) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z127, Q9JMI4 | Gene names | Slc7a5, Lat1 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC). | |||||
|
XCT_HUMAN
|
||||||
| NC score | 0.071797 (rank : 32) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPY5 | Gene names | SLC7A11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT) (Calcium channel blocker resistance protein CCBR1). | |||||
|
G137B_HUMAN
|
||||||
| NC score | 0.049825 (rank : 33) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
LIN1_HUMAN
|
||||||
| NC score | 0.018014 (rank : 34) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.017030 (rank : 35) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.016855 (rank : 36) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CT098_HUMAN
|
||||||
| NC score | 0.011676 (rank : 37) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZP1, Q6FII5, Q9NUD3 | Gene names | C20orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf98. | |||||
|
NBEA_HUMAN
|
||||||
| NC score | 0.011657 (rank : 38) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFP9, Q9HCM8, Q9NSU1, Q9NW98, Q9Y6J1 | Gene names | NBEA, BCL8B, KIAA1544, LYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2) (Protein BCL8B). | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.011549 (rank : 39) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
RHBD4_HUMAN
|
||||||
| NC score | 0.010318 (rank : 40) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
GPR84_HUMAN
|
||||||
| NC score | 0.009460 (rank : 41) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQS5 | Gene names | GPR84, EX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 84 (Inflammation-related G-protein coupled receptor EX33). | |||||
|
GP149_MOUSE
|
||||||
| NC score | 0.009299 (rank : 42) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UVY1, Q3TMB1, Q80T52, Q8BXA3, Q8BZC0 | Gene names | Gpr149, Pgr10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
RHBD4_MOUSE
|
||||||
| NC score | 0.008354 (rank : 43) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.007585 (rank : 44) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EDG1_MOUSE
|
||||||
| NC score | 0.005718 (rank : 45) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08530, Q9R235 | Gene names | Edg1, Lpb1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1) (Lysophospholipid receptor B1). | |||||
|
F13B_HUMAN
|
||||||
| NC score | 0.004929 (rank : 46) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
F13B_MOUSE
|
||||||
| NC score | 0.004348 (rank : 47) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
IRK8_HUMAN
|
||||||
| NC score | 0.002842 (rank : 48) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15842, O00657 | Gene names | KCNJ8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 8 (Potassium channel, inwardly rectifying subfamily J member 8) (Inwardly rectifier K(+) channel Kir6.1) (uKATP-1). | |||||
|
O1052_MOUSE
|
||||||
| NC score | 0.001679 (rank : 49) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VGR8 | Gene names | Olfr1052, Mor172-1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1052 (Olfactory receptor 172-1). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.000866 (rank : 50) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
OR8U1_HUMAN
|
||||||
| NC score | -0.000058 (rank : 51) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NH10 | Gene names | OR8U1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 8U1. | |||||
|
OL998_MOUSE
|
||||||
| NC score | -0.000066 (rank : 52) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 648 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VF76 | Gene names | Olfr998, Mor175-5 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 998 (Olfactory receptor 175-5). | |||||
|
OR8B8_HUMAN
|
||||||
| NC score | -0.000149 (rank : 53) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15620, Q96RC8 | Gene names | OR8B8 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 8B8 (Olfactory receptor TPCR85) (Olfactory-like receptor JCG8). | |||||
|
O1044_MOUSE
|
||||||
| NC score | -0.000150 (rank : 54) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VGR9 | Gene names | Olfr1044, Mor185-4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1044 (Olfactory receptor 185-4). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | -0.000310 (rank : 55) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
5HT1B_MOUSE
|
||||||
| NC score | -0.001053 (rank : 56) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28334 | Gene names | Htr1b, 5ht1b | |||
|
Domain Architecture |

|
|||||
| Description | 5-hydroxytryptamine 1B receptor (5-HT-1B) (Serotonin receptor 1B) (5- HT1B). | |||||
|
KLF8_HUMAN
|
||||||
| NC score | -0.002131 (rank : 57) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95600, Q59GV5, Q5HYQ5, Q5JXP7, Q6MZJ7, Q9UGC4 | Gene names | KLF8, BKLF3, ZNF741 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 8 (Basic krueppel-like factor 3) (Zinc finger protein 741). | |||||
|
ZFP95_MOUSE
|
||||||
| NC score | -0.002844 (rank : 58) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1D8 | Gene names | Zfp95 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 (Zfp-95). | |||||