Please be patient as the page loads
|
HTRA2_HUMAN
|
||||||
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HTRA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_MOUSE
|
||||||
| θ value | 7.71569e-185 (rank : 2) | NC score | 0.997314 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 1.06975e-93 (rank : 3) | NC score | 0.907065 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 5.86991e-92 (rank : 4) | NC score | 0.902841 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 1.88829e-90 (rank : 5) | NC score | 0.868504 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 1.03614e-88 (rank : 6) | NC score | 0.871812 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 1.49966e-79 (rank : 7) | NC score | 0.910634 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 8) | NC score | 0.151030 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.149079 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.044800 (rank : 73) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.049461 (rank : 70) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
PDLI3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.052437 (rank : 65) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q53GG5, O43590, O60439, O60440, Q8N6Y6, Q9BVP4 | Gene names | PDLIM3, ALP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.103750 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
BHLH4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.025331 (rank : 77) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
PDLI2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.052918 (rank : 63) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R1G6 | Gene names | Pdlim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.031010 (rank : 75) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
PDLI3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.046466 (rank : 72) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70209 | Gene names | Pdlim3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.006183 (rank : 83) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.030249 (rank : 76) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.102582 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.048989 (rank : 71) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
PDLI2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.042595 (rank : 74) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.021755 (rank : 78) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
CO5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.008330 (rank : 82) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.084996 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.016806 (rank : 79) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CREG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.015607 (rank : 80) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUH2, Q86X03, Q8N540, Q8N9E3 | Gene names | CREG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CREG2 precursor. | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.009451 (rank : 81) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.052766 (rank : 64) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
SEM3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.002634 (rank : 84) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.079124 (rank : 40) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
CTGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.076179 (rank : 42) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |
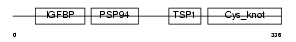
|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
CTGF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.054782 (rank : 58) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |
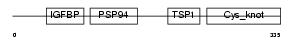
|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
CYR61_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.089776 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
CYR61_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.089940 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
ESM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.195096 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
ESM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.200043 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |
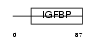
|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.051601 (rank : 66) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050240 (rank : 69) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
GRASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.050636 (rank : 67) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.057972 (rank : 53) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
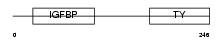
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.094363 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.085325 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
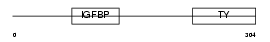
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.073147 (rank : 45) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.109626 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.078539 (rank : 41) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.068316 (rank : 48) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
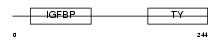
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.064343 (rank : 51) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
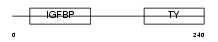
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.081663 (rank : 39) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.081804 (rank : 38) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.266865 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
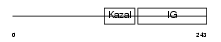
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.260231 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
IPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.069814 (rank : 47) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.100268 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.211163 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.200632 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.141390 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.220881 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.252975 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.222357 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.274056 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.128137 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.065420 (rank : 50) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.105883 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.086348 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.107028 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.059949 (rank : 52) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.121863 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.075536 (rank : 43) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.072360 (rank : 46) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NOV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.083100 (rank : 37) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
NOV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.075418 (rank : 44) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.084496 (rank : 36) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
RHPN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.057101 (rank : 55) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
RHPN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.053990 (rank : 59) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
TX1B3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.055664 (rank : 56) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.055068 (rank : 57) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052961 (rank : 62) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.050617 (rank : 68) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.053595 (rank : 61) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.053721 (rank : 60) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
WISP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.087846 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
WISP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057321 (rank : 54) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |
|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
WISP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.067841 (rank : 49) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
HTRA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_MOUSE
|
||||||
| NC score | 0.997314 (rank : 2) | θ value | 7.71569e-185 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.910634 (rank : 3) | θ value | 1.49966e-79 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.907065 (rank : 4) | θ value | 1.06975e-93 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.902841 (rank : 5) | θ value | 5.86991e-92 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.871812 (rank : 6) | θ value | 1.03614e-88 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.868504 (rank : 7) | θ value | 1.88829e-90 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.274056 (rank : 8) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.266865 (rank : 9) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.260231 (rank : 10) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.252975 (rank : 11) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.222357 (rank : 12) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.220881 (rank : 13) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.211163 (rank : 14) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.200632 (rank : 15) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
ESM1_MOUSE
|
||||||
| NC score | 0.200043 (rank : 16) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.195096 (rank : 17) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.151030 (rank : 18) | θ value | 0.00175202 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.149079 (rank : 19) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.141390 (rank : 20) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.128137 (rank : 21) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.121863 (rank : 22) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.109626 (rank : 23) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.107028 (rank : 24) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.105883 (rank : 25) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.103750 (rank : 26) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.102582 (rank : 27) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.100268 (rank : 28) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.094363 (rank : 29) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
CYR61_MOUSE
|
||||||
| NC score | 0.089940 (rank : 30) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
CYR61_HUMAN
|
||||||
| NC score | 0.089776 (rank : 31) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
WISP2_HUMAN
|
||||||
| NC score | 0.087846 (rank : 32) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.086348 (rank : 33) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.085325 (rank : 34) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.084996 (rank : 35) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.084496 (rank : 36) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.083100 (rank : 37) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.081804 (rank : 38) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.081663 (rank : 39) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.079124 (rank : 40) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.078539 (rank : 41) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
CTGF_HUMAN
|
||||||
| NC score | 0.076179 (rank : 42) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.075536 (rank : 43) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.075418 (rank : 44) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.073147 (rank : 45) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.072360 (rank : 46) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.069814 (rank : 47) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.068316 (rank : 48) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
WISP3_HUMAN
|
||||||
| NC score | 0.067841 (rank : 49) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.065420 (rank : 50) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.064343 (rank : 51) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.059949 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.057972 (rank : 53) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
WISP2_MOUSE
|
||||||
| NC score | 0.057321 (rank : 54) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
RHPN1_HUMAN
|
||||||
| NC score | 0.057101 (rank : 55) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
TX1B3_HUMAN
|
||||||
| NC score | 0.055664 (rank : 56) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| NC score | 0.055068 (rank : 57) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
CTGF_MOUSE
|
||||||
| NC score | 0.054782 (rank : 58) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
RHPN1_MOUSE
|
||||||
| NC score | 0.053990 (rank : 59) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.053721 (rank : 60) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.053595 (rank : 61) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.052961 (rank : 62) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
PDLI2_MOUSE
|
||||||
| NC score | 0.052918 (rank : 63) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R1G6 | Gene names | Pdlim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.052766 (rank : 64) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
PDLI3_HUMAN
|
||||||
| NC score | 0.052437 (rank : 65) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q53GG5, O43590, O60439, O60440, Q8N6Y6, Q9BVP4 | Gene names | PDLIM3, ALP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.051601 (rank : 66) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
GRASP_HUMAN
|
||||||
| NC score | 0.050636 (rank : 67) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.050617 (rank : 68) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.050240 (rank : 69) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.049461 (rank : 70) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.048989 (rank : 71) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
PDLI3_MOUSE
|
||||||
| NC score | 0.046466 (rank : 72) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70209 | Gene names | Pdlim3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.044800 (rank : 73) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
PDLI2_HUMAN
|
||||||
| NC score | 0.042595 (rank : 74) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.031010 (rank : 75) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.030249 (rank : 76) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
BHLH4_HUMAN
|
||||||
| NC score | 0.025331 (rank : 77) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.021755 (rank : 78) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.016806 (rank : 79) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CREG2_HUMAN
|
||||||
| NC score | 0.015607 (rank : 80) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUH2, Q86X03, Q8N540, Q8N9E3 | Gene names | CREG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CREG2 precursor. | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.009451 (rank : 81) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
CO5_MOUSE
|
||||||
| NC score | 0.008330 (rank : 82) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.006183 (rank : 83) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
SEM3B_HUMAN
|
||||||
| NC score | 0.002634 (rank : 84) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||