Please be patient as the page loads
|
CHLE_HUMAN
|
||||||
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHLE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998874 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
ACES_HUMAN
|
||||||
| θ value | 4.52334e-185 (rank : 3) | NC score | 0.993595 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_MOUSE
|
||||||
| θ value | 3.96267e-181 (rank : 4) | NC score | 0.993998 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
NLGNY_HUMAN
|
||||||
| θ value | 3.11973e-85 (rank : 5) | NC score | 0.946700 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGNX_HUMAN
|
||||||
| θ value | 1.18549e-84 (rank : 6) | NC score | 0.946507 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | 4.21644e-82 (rank : 7) | NC score | 0.948263 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_HUMAN
|
||||||
| θ value | 3.56943e-81 (rank : 8) | NC score | 0.948154 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | 5.69867e-79 (rank : 9) | NC score | 0.945462 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN3_HUMAN
|
||||||
| θ value | 2.39424e-77 (rank : 10) | NC score | 0.944702 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |
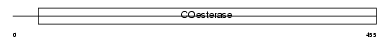
|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN1_HUMAN
|
||||||
| θ value | 5.33382e-77 (rank : 11) | NC score | 0.944050 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_MOUSE
|
||||||
| θ value | 7.70199e-76 (rank : 12) | NC score | 0.943954 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |
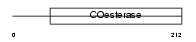
|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
CEL_MOUSE
|
||||||
| θ value | 1.27248e-70 (rank : 13) | NC score | 0.963990 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST2_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 14) | NC score | 0.961375 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.40689e-69 (rank : 15) | NC score | 0.729242 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CES3_MOUSE
|
||||||
| θ value | 1.45591e-66 (rank : 16) | NC score | 0.943389 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
EST1_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 17) | NC score | 0.943416 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
ESTN_MOUSE
|
||||||
| θ value | 7.47731e-63 (rank : 18) | NC score | 0.943964 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST22_MOUSE
|
||||||
| θ value | 1.66577e-62 (rank : 19) | NC score | 0.945064 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
EST1_MOUSE
|
||||||
| θ value | 1.19377e-60 (rank : 20) | NC score | 0.940477 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
EST31_MOUSE
|
||||||
| θ value | 1.19377e-60 (rank : 21) | NC score | 0.955216 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 1.45929e-58 (rank : 22) | NC score | 0.840715 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 23) | NC score | 0.828352 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
AAAD_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 24) | NC score | 0.291407 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.075892 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
LIPS_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.078342 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |
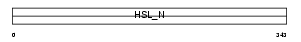
|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
TIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.019844 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TB96, Q96SR4, Q9BRE2, Q9H2V9 | Gene names | ITFG1, TIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunomodulatory protein precursor (Protein TIP) (Integrin alpha FG-GAP repeat-containing protein 1). | |||||
|
SEM4G_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.004822 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
AAAD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.171719 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.007301 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
TSN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.006505 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14817 | Gene names | TSPAN4, NAG2, TM4SF7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-4 (Tspan-4) (Transmembrane 4 superfamily member 7) (Novel antigen 2) (NAG-2). | |||||
|
CAYP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.011159 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.010482 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.003087 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
TSN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.004743 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCK3 | Gene names | Tspan4, Tm4sf7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-4 (Tspan-4) (Transmembrane 4 superfamily member 7). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.074359 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
CHLE_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_MOUSE
|
||||||
| NC score | 0.998874 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
ACES_MOUSE
|
||||||
| NC score | 0.993998 (rank : 3) | θ value | 3.96267e-181 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_HUMAN
|
||||||
| NC score | 0.993595 (rank : 4) | θ value | 4.52334e-185 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
CEL_MOUSE
|
||||||
| NC score | 0.963990 (rank : 5) | θ value | 1.27248e-70 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST2_HUMAN
|
||||||
| NC score | 0.961375 (rank : 6) | θ value | 4.83544e-70 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
EST31_MOUSE
|
||||||
| NC score | 0.955216 (rank : 7) | θ value | 1.19377e-60 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.948263 (rank : 8) | θ value | 4.21644e-82 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_HUMAN
|
||||||
| NC score | 0.948154 (rank : 9) | θ value | 3.56943e-81 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGNY_HUMAN
|
||||||
| NC score | 0.946700 (rank : 10) | θ value | 3.11973e-85 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGNX_HUMAN
|
||||||
| NC score | 0.946507 (rank : 11) | θ value | 1.18549e-84 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.945462 (rank : 12) | θ value | 5.69867e-79 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
EST22_MOUSE
|
||||||
| NC score | 0.945064 (rank : 13) | θ value | 1.66577e-62 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
NLGN3_HUMAN
|
||||||
| NC score | 0.944702 (rank : 14) | θ value | 2.39424e-77 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN1_HUMAN
|
||||||
| NC score | 0.944050 (rank : 15) | θ value | 5.33382e-77 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
ESTN_MOUSE
|
||||||
| NC score | 0.943964 (rank : 16) | θ value | 7.47731e-63 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
NLGN1_MOUSE
|
||||||
| NC score | 0.943954 (rank : 17) | θ value | 7.70199e-76 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
EST1_HUMAN
|
||||||
| NC score | 0.943416 (rank : 18) | θ value | 5.17823e-64 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
CES3_MOUSE
|
||||||
| NC score | 0.943389 (rank : 19) | θ value | 1.45591e-66 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
EST1_MOUSE
|
||||||
| NC score | 0.940477 (rank : 20) | θ value | 1.19377e-60 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.840715 (rank : 21) | θ value | 1.45929e-58 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.828352 (rank : 22) | θ value | 1.08223e-53 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.729242 (rank : 23) | θ value | 1.40689e-69 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
AAAD_HUMAN
|
||||||
| NC score | 0.291407 (rank : 24) | θ value | 0.00020696 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| NC score | 0.171719 (rank : 25) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
LIPS_MOUSE
|
||||||
| NC score | 0.078342 (rank : 26) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.075892 (rank : 27) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.074359 (rank : 28) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
TIP_HUMAN
|
||||||
| NC score | 0.019844 (rank : 29) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TB96, Q96SR4, Q9BRE2, Q9H2V9 | Gene names | ITFG1, TIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunomodulatory protein precursor (Protein TIP) (Integrin alpha FG-GAP repeat-containing protein 1). | |||||
|
CAYP2_MOUSE
|
||||||
| NC score | 0.011159 (rank : 30) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.010482 (rank : 31) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.007301 (rank : 32) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
TSN4_HUMAN
|
||||||
| NC score | 0.006505 (rank : 33) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14817 | Gene names | TSPAN4, NAG2, TM4SF7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-4 (Tspan-4) (Transmembrane 4 superfamily member 7) (Novel antigen 2) (NAG-2). | |||||
|
SEM4G_HUMAN
|
||||||
| NC score | 0.004822 (rank : 34) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
TSN4_MOUSE
|
||||||
| NC score | 0.004743 (rank : 35) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCK3 | Gene names | Tspan4, Tm4sf7 | |||
|
Domain Architecture |

|
|||||
| Description | Tetraspanin-4 (Tspan-4) (Transmembrane 4 superfamily member 7). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.003087 (rank : 36) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||