Please be patient as the page loads
|
AAAD_HUMAN
|
||||||
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AAAD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| θ value | 3.60074e-166 (rank : 2) | NC score | 0.971699 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 3) | NC score | 0.320113 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
LIPS_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 4) | NC score | 0.357416 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |
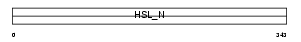
|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
CHLE_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 5) | NC score | 0.281899 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.291407 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CEL_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 7) | NC score | 0.276634 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 8) | NC score | 0.211375 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ACES_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 9) | NC score | 0.284971 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
EST1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 10) | NC score | 0.287958 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
EST31_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 11) | NC score | 0.295924 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
EST2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 12) | NC score | 0.274503 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
CES3_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 13) | NC score | 0.282386 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
ACES_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 14) | NC score | 0.277583 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
NLGN1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.231323 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.231404 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |
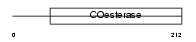
|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
EST22_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.278840 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
ESTN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.271259 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.263731 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
GSTT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.030014 (rank : 30) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30711, O00226, Q969K8, Q96IY3 | Gene names | GSTT1 | |||
|
Domain Architecture |
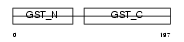
|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1) (Glutathione transferase T1-1). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.011077 (rank : 31) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.011070 (rank : 32) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
AFMID_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.059320 (rank : 28) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q63HM1 | Gene names | AFMID | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable arylformamidase (EC 3.5.1.9) (Kynurenine formamidase) (KF). | |||||
|
AFMID_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.052996 (rank : 29) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4H1, Q80XP3, Q8R1K6 | Gene names | Afmid | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable arylformamidase (EC 3.5.1.9) (Kynurenine formamidase) (KF). | |||||
|
NLGN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.208575 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.208508 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.205360 (rank : 25) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |
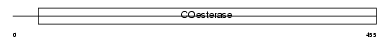
|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.206002 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGNX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.206764 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGNY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.206796 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
THYG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.178834 (rank : 27) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.180217 (rank : 26) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
AAAD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| NC score | 0.971699 (rank : 2) | θ value | 3.60074e-166 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
LIPS_MOUSE
|
||||||
| NC score | 0.357416 (rank : 3) | θ value | 7.59969e-07 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.320113 (rank : 4) | θ value | 1.17247e-07 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
EST31_MOUSE
|
||||||
| NC score | 0.295924 (rank : 5) | θ value | 0.00134147 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
CHLE_HUMAN
|
||||||
| NC score | 0.291407 (rank : 6) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
EST1_HUMAN
|
||||||
| NC score | 0.287958 (rank : 7) | θ value | 0.00134147 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
ACES_MOUSE
|
||||||
| NC score | 0.284971 (rank : 8) | θ value | 0.00134147 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
CES3_MOUSE
|
||||||
| NC score | 0.282386 (rank : 9) | θ value | 0.00390308 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
CHLE_MOUSE
|
||||||
| NC score | 0.281899 (rank : 10) | θ value | 9.29e-05 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
EST22_MOUSE
|
||||||
| NC score | 0.278840 (rank : 11) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
ACES_HUMAN
|
||||||
| NC score | 0.277583 (rank : 12) | θ value | 0.00665767 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
CEL_MOUSE
|
||||||
| NC score | 0.276634 (rank : 13) | θ value | 0.000602161 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST2_HUMAN
|
||||||
| NC score | 0.274503 (rank : 14) | θ value | 0.00298849 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
ESTN_MOUSE
|
||||||
| NC score | 0.271259 (rank : 15) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST1_MOUSE
|
||||||
| NC score | 0.263731 (rank : 16) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
NLGN1_MOUSE
|
||||||
| NC score | 0.231404 (rank : 17) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_HUMAN
|
||||||
| NC score | 0.231323 (rank : 18) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.211375 (rank : 19) | θ value | 0.000786445 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NLGN2_HUMAN
|
||||||
| NC score | 0.208575 (rank : 20) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.208508 (rank : 21) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGNY_HUMAN
|
||||||
| NC score | 0.206796 (rank : 22) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGNX_HUMAN
|
||||||
| NC score | 0.206764 (rank : 23) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.206002 (rank : 24) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN3_HUMAN
|
||||||
| NC score | 0.205360 (rank : 25) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.180217 (rank : 26) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.178834 (rank : 27) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
AFMID_HUMAN
|
||||||
| NC score | 0.059320 (rank : 28) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q63HM1 | Gene names | AFMID | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable arylformamidase (EC 3.5.1.9) (Kynurenine formamidase) (KF). | |||||
|
AFMID_MOUSE
|
||||||
| NC score | 0.052996 (rank : 29) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4H1, Q80XP3, Q8R1K6 | Gene names | Afmid | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable arylformamidase (EC 3.5.1.9) (Kynurenine formamidase) (KF). | |||||
|
GSTT1_HUMAN
|
||||||
| NC score | 0.030014 (rank : 30) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30711, O00226, Q969K8, Q96IY3 | Gene names | GSTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1) (Glutathione transferase T1-1). | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.011077 (rank : 31) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.011070 (rank : 32) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||