Please be patient as the page loads
|
BIG2_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BIG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992959 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
BIG2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
GBF1_HUMAN
|
||||||
| θ value | 4.10295e-61 (rank : 3) | NC score | 0.868836 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
CYH1_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 4) | NC score | 0.819373 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
|
|||||
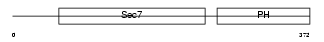
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 1.61718e-49 (rank : 5) | NC score | 0.817051 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 3.37202e-47 (rank : 6) | NC score | 0.820080 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 5.7518e-47 (rank : 7) | NC score | 0.814099 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 8) | NC score | 0.819934 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 1.56636e-44 (rank : 9) | NC score | 0.819922 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 10) | NC score | 0.819422 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 11) | NC score | 0.818462 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
IQEC1_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 12) | NC score | 0.801806 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R0S2, Q3TZC3, Q5DU15 | Gene names | Iqsec1, Kiaa0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1. | |||||
|
IQEC1_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 13) | NC score | 0.799381 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
IQEC3_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 14) | NC score | 0.805995 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 15) | NC score | 0.797991 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 16) | NC score | 0.755825 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 1.6813e-30 (rank : 17) | NC score | 0.755913 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PSD4_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 18) | NC score | 0.687635 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
PSD3_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 19) | NC score | 0.726307 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
FBX8_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 20) | NC score | 0.758627 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QZN3 | Gene names | Fbxo8, Fbx8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 8. | |||||
|
FBX8_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 21) | NC score | 0.756173 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NRD0, Q6UWN4, Q9NRP5, Q9UKC4 | Gene names | FBXO8, FBS, FBX8 | |||
|
Domain Architecture |
|
|||||
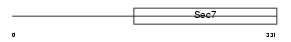
| Description | F-box only protein 8 (F-box/SEC7 protein FBS). | |||||
|
PSD4_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 22) | NC score | 0.691203 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.029284 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.018752 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.018824 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
PEX14_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.032533 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |
|
|||||
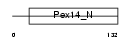
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.009922 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.015295 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
|
|||||
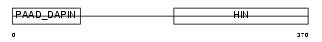
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.011872 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.011589 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.000568 (rank : 101) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.011309 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.004140 (rank : 97) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
GP73_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.013543 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
ADAM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.003598 (rank : 98) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60718, Q60814, Q9D4G3, Q9QWJ0 | Gene names | Adam2, Ftnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 2 precursor (A disintegrin and metalloproteinase domain 2) (Fertilin subunit beta) (PH-30) (PH30) (PH30-beta). | |||||
|
ANR47_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.004705 (rank : 93) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.008923 (rank : 82) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.008543 (rank : 84) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.008255 (rank : 86) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.009707 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.012290 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CD5R1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.016891 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CD5R1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.016815 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61809, Q62938 | Gene names | Cdk5r1, Cdk5r, Nck5a | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.005160 (rank : 90) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
RASA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.036135 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.008749 (rank : 83) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.008298 (rank : 85) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
E2F1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.012714 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61501, Q80VZ3 | Gene names | E2f1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.004182 (rank : 96) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.007330 (rank : 87) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
LGR6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.001411 (rank : 100) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBX8, Q5T509, Q5T512, Q6UY15, Q86VU0, Q96K69, Q9BYD7 | Gene names | LGR6 | |||
|
Domain Architecture |
|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 6 (VTS20631). | |||||
|
NOCT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.010804 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35710, Q9QZG9 | Gene names | Ccrn4l, Ccr4, Noc | |||
|
Domain Architecture |
|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | -0.001580 (rank : 104) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
ZFX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | -0.001455 (rank : 103) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17010, O43668, Q8WYJ8 | Gene names | ZFX | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
ZFX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | -0.001448 (rank : 102) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17012, P17011 | Gene names | Zfx | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
CC062_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.011000 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
DDX11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.006770 (rank : 88) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6AXC6 | Gene names | Ddx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11). | |||||
|
ENOA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.004924 (rank : 92) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06733, P22712, Q16704, Q4TUS4, Q658M5, Q6GMP2, Q71V37, Q7Z3V6, Q8WU71, Q9UM55 | Gene names | ENO1, ENO1L1, MBPB1, MPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-enolase (EC 4.2.1.11) (2-phospho-D-glycerate hydro-lyase) (Non- neural enolase) (NNE) (Enolase 1) (Phosphopyruvate hydratase) (C-myc promoter-binding protein) (MBP-1) (MPB-1) (Plasminogen-binding protein). | |||||
|
GEMI4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.011873 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P57678, Q9NZS7, Q9UG32, Q9Y4Q2 | Gene names | GEMIN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Component of gems 4 (Gemin-4) (p97). | |||||
|
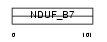
NDUB7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.013599 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17568, Q9UI16 | Gene names | NDUFB7 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B18 subunit) (Complex I-B18) (CI-B18) (Cell adhesion protein SQM1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.004704 (rank : 94) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.005227 (rank : 89) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.003277 (rank : 99) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.005144 (rank : 91) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.004474 (rank : 95) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
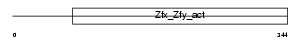
ZFA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | -0.001610 (rank : 105) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23607 | Gene names | Zfa | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger autosomal protein. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.107587 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.111468 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.114396 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
ANLN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.095331 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CENB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.065749 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.060713 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.184954 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.096834 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.096743 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
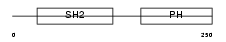
DAPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.109990 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
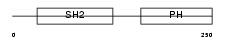
DAPP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.115678 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DYN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052593 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.052557 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.053145 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056023 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053405 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.053391 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.133092 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.132628 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.103277 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.117065 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.110635 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.120365 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.093286 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.130094 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.124825 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051305 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |
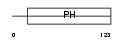
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PKHB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055970 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96CS7, Q86W37, Q9BV75, Q9NWK1 | Gene names | PLEKHB2, EVT2 | |||
|
Domain Architecture |
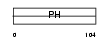
|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
PKHB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.054844 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZC7, Q3UDH6, Q8C4I4, Q8CA27 | Gene names | Plekhb2, Evt2 | |||
|
Domain Architecture |
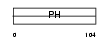
|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.099170 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
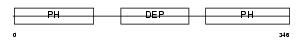
|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.100852 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
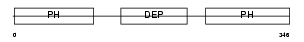
|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.128758 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.132013 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
RASA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.079703 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.074191 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.077105 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SWP70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.070219 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.061378 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.051135 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
BIG2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
BIG1_HUMAN
|
||||||
| NC score | 0.992959 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y6D6, Q9NV46, Q9UFV2, Q9UNL0 | Gene names | ARFGEF1, ARFGEP1, BIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (Brefeldin A-inhibited GEP 1) (p200 ARF-GEP1) (p200 ARF guanine nucleotide exchange factor). | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.868836 (rank : 3) | θ value | 4.10295e-61 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.820080 (rank : 4) | θ value | 3.37202e-47 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.819934 (rank : 5) | θ value | 9.18288e-45 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.819922 (rank : 6) | θ value | 1.56636e-44 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.819422 (rank : 7) | θ value | 3.48949e-44 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.819373 (rank : 8) | θ value | 1.23823e-49 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.818462 (rank : 9) | θ value | 6.58091e-43 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.817051 (rank : 10) | θ value | 1.61718e-49 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.814099 (rank : 11) | θ value | 5.7518e-47 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
IQEC3_HUMAN
|
||||||
| NC score | 0.805995 (rank : 12) | θ value | 4.42448e-31 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPP2, Q8TB43 | Gene names | IQSEC3, KIAA1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
IQEC1_MOUSE
|
||||||
| NC score | 0.801806 (rank : 13) | θ value | 4.00176e-32 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R0S2, Q3TZC3, Q5DU15 | Gene names | Iqsec1, Kiaa0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1. | |||||
|
IQEC1_HUMAN
|
||||||
| NC score | 0.799381 (rank : 14) | θ value | 5.22648e-32 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.797991 (rank : 15) | θ value | 4.42448e-31 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
FBX8_MOUSE
|
||||||
| NC score | 0.758627 (rank : 16) | θ value | 1.16975e-15 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QZN3 | Gene names | Fbxo8, Fbx8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 8. | |||||
|
FBX8_HUMAN
|
||||||
| NC score | 0.756173 (rank : 17) | θ value | 1.99529e-15 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NRD0, Q6UWN4, Q9NRP5, Q9UKC4 | Gene names | FBXO8, FBS, FBX8 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 8 (F-box/SEC7 protein FBS). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.755913 (rank : 18) | θ value | 1.6813e-30 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.755825 (rank : 19) | θ value | 1.28732e-30 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PSD3_HUMAN
|
||||||
| NC score | 0.726307 (rank : 20) | θ value | 6.85773e-16 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYI0, Q9Y2F1 | Gene names | PSD3, EFA6R, HCA67, KIAA0942 | |||
|
Domain Architecture |

|
|||||
| Description | PH and SEC7 domain-containing protein 3 (Pleckstrin homology and SEC7 domain-containing protein 3) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6) (Hepatocellular carcinoma- associated antigen 67). | |||||
|
PSD4_MOUSE
|
||||||
| NC score | 0.691203 (rank : 21) | θ value | 2.60593e-15 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.687635 (rank : 22) | θ value | 1.63225e-17 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.184954 (rank : 23) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.133092 (rank : 24) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.132628 (rank : 25) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.132013 (rank : 26) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.130094 (rank : 27) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.128758 (rank : 28) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.124825 (rank : 29) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.120365 (rank : 30) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.117065 (rank : 31) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.115678 (rank : 32) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.114396 (rank : 33) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.111468 (rank : 34) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.110635 (rank : 35) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.109990 (rank : 36) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.107587 (rank : 37) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.103277 (rank : 38) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.100852 (rank : 39) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.099170 (rank : 40) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.096834 (rank : 41) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.096743 (rank : 42) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.095331 (rank : 43) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.093286 (rank : 44) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
RASA1_HUMAN
|
||||||
| NC score | 0.079703 (rank : 45) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.077105 (rank : 46) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.074191 (rank : 47) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.070219 (rank : 48) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.065749 (rank : 49) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.061378 (rank : 50) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.060713 (rank : 51) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.056023 (rank : 52) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
PKHB2_HUMAN
|
||||||
| NC score | 0.055970 (rank : 53) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96CS7, Q86W37, Q9BV75, Q9NWK1 | Gene names | PLEKHB2, EVT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
PKHB2_MOUSE
|
||||||
| NC score | 0.054844 (rank : 54) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZC7, Q3UDH6, Q8C4I4, Q8CA27 | Gene names | Plekhb2, Evt2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 2 (Evectin-2). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.053405 (rank : 55) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.053391 (rank : 56) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.053145 (rank : 57) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DYN2_HUMAN
|
||||||
| NC score | 0.052593 (rank : 58) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| NC score | 0.052557 (rank : 59) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
PKHB1_HUMAN
|
||||||
| NC score | 0.051305 (rank : 60) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.051135 (rank : 61) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.036135 (rank : 62) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
PEX14_MOUSE
|
||||||
| NC score | 0.032533 (rank : 63) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.029284 (rank : 64) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.018824 (rank : 65) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.018752 (rank : 66) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
CD5R1_HUMAN
|
||||||
| NC score | 0.016891 (rank : 67) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CD5R1_MOUSE
|
||||||
| NC score | 0.016815 (rank : 68) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61809, Q62938 | Gene names | Cdk5r1, Cdk5r, Nck5a | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.015295 (rank : 69) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
NDUB7_HUMAN
|
||||||
| NC score | 0.013599 (rank : 70) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17568, Q9UI16 | Gene names | NDUFB7 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B18 subunit) (Complex I-B18) (CI-B18) (Cell adhesion protein SQM1). | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.013543 (rank : 71) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
E2F1_MOUSE
|
||||||
| NC score | 0.012714 (rank : 72) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61501, Q80VZ3 | Gene names | E2f1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.012290 (rank : 73) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
GEMI4_HUMAN
|
||||||
| NC score | 0.011873 (rank : 74) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P57678, Q9NZS7, Q9UG32, Q9Y4Q2 | Gene names | GEMIN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Component of gems 4 (Gemin-4) (p97). | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.011872 (rank : 75) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.011589 (rank : 76) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.011309 (rank : 77) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
CC062_HUMAN
|
||||||
| NC score | 0.011000 (rank : 78) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
NOCT_MOUSE
|
||||||
| NC score | 0.010804 (rank : 79) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35710, Q9QZG9 | Gene names | Ccrn4l, Ccr4, Noc | |||
|
Domain Architecture |
|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.009922 (rank : 80) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.009707 (rank : 81) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.008923 (rank : 82) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.008749 (rank : 83) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.008543 (rank : 84) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.008298 (rank : 85) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.008255 (rank : 86) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.007330 (rank : 87) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
DDX11_MOUSE
|
||||||
| NC score | 0.006770 (rank : 88) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6AXC6 | Gene names | Ddx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.005227 (rank : 89) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.005160 (rank : 90) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.005144 (rank : 91) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
ENOA_HUMAN
|
||||||
| NC score | 0.004924 (rank : 92) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06733, P22712, Q16704, Q4TUS4, Q658M5, Q6GMP2, Q71V37, Q7Z3V6, Q8WU71, Q9UM55 | Gene names | ENO1, ENO1L1, MBPB1, MPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-enolase (EC 4.2.1.11) (2-phospho-D-glycerate hydro-lyase) (Non- neural enolase) (NNE) (Enolase 1) (Phosphopyruvate hydratase) (C-myc promoter-binding protein) (MBP-1) (MPB-1) (Plasminogen-binding protein). | |||||
|
ANR47_MOUSE
|
||||||
| NC score | 0.004705 (rank : 93) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.004704 (rank : 94) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.004474 (rank : 95) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.004182 (rank : 96) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.004140 (rank : 97) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
ADAM2_MOUSE
|
||||||
| NC score | 0.003598 (rank : 98) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60718, Q60814, Q9D4G3, Q9QWJ0 | Gene names | Adam2, Ftnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 2 precursor (A disintegrin and metalloproteinase domain 2) (Fertilin subunit beta) (PH-30) (PH30) (PH30-beta). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.003277 (rank : 99) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
LGR6_HUMAN
|
||||||
| NC score | 0.001411 (rank : 100) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBX8, Q5T509, Q5T512, Q6UY15, Q86VU0, Q96K69, Q9BYD7 | Gene names | LGR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 6 (VTS20631). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.000568 (rank : 101) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
ZFX_MOUSE
|
||||||
| NC score | -0.001448 (rank : 102) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17012, P17011 | Gene names | Zfx | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
ZFX_HUMAN
|
||||||
| NC score | -0.001455 (rank : 103) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17010, O43668, Q8WYJ8 | Gene names | ZFX | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger X-chromosomal protein. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | -0.001580 (rank : 104) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
ZFA_MOUSE
|
||||||
| NC score | -0.001610 (rank : 105) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23607 | Gene names | Zfa | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger autosomal protein. | |||||