Please be patient as the page loads
|
NOCT_MOUSE
|
||||||
| SwissProt Accessions | O35710, Q9QZG9 | Gene names | Ccrn4l, Ccr4, Noc | |||
|
Domain Architecture |
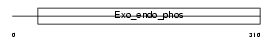
|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NOCT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996179 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UK39, Q9HD93, Q9HD94, Q9HD95 | Gene names | CCRN4L, CCR4, NOC | |||
|
Domain Architecture |
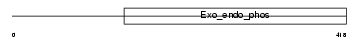
|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
|
NOCT_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35710, Q9QZG9 | Gene names | Ccrn4l, Ccr4, Noc | |||
|
Domain Architecture |
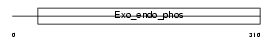
|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
|
CNOT6_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 3) | NC score | 0.357457 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3P5, Q5NCL3, Q80V15 | Gene names | Cnot6, Ccr4, Kiaa1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
CNOT6_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 4) | NC score | 0.372832 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULM6 | Gene names | CNOT6, CCR4, KIAA1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
ANGE1_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 5) | NC score | 0.610463 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNK9, O94859 | Gene names | ANGEL1, KIAA0759 | |||
|
Domain Architecture |
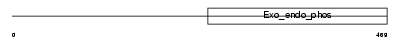
|
|||||
| Description | Angel homolog 1. | |||||
|
TTRAP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.200460 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95551, Q2TBE2, Q5JYM0, Q7Z6U5, Q9NUK5, Q9NYY9 | Gene names | TTRAP, EAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF and TNF receptor-associated protein (ETS1-associated protein 2) (ETS1-associated protein II) (EAPII). | |||||
|
TTRAP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.159942 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJX7 | Gene names | Ttrap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF and TNF receptor-associated protein. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.024052 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PTN22_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.013832 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
CSTFT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.016980 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
CSTFT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.016472 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.013309 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
CJ006_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.029199 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IX21, Q5W0L8, Q9NPE8 | Gene names | C10orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6. | |||||
|
BIG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.010804 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.001247 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.017572 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
B4GN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.012270 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00973 | Gene names | B4GALNT1, GALGT, SIAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4 N-acetylgalactosaminyltransferase 1 (EC 2.4.1.92) ((N- acetylneuraminyl)-galactosylglucosylceramide) (GM2/GD2 synthase) (GalNAc-T). | |||||
|
NOCT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35710, Q9QZG9 | Gene names | Ccrn4l, Ccr4, Noc | |||
|
Domain Architecture |
|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
|
NOCT_HUMAN
|
||||||
| NC score | 0.996179 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UK39, Q9HD93, Q9HD94, Q9HD95 | Gene names | CCRN4L, CCR4, NOC | |||
|
Domain Architecture |

|
|||||
| Description | Nocturnin (CCR4 protein homolog). | |||||
|
ANGE1_HUMAN
|
||||||
| NC score | 0.610463 (rank : 3) | θ value | 1.52774e-15 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNK9, O94859 | Gene names | ANGEL1, KIAA0759 | |||
|
Domain Architecture |

|
|||||
| Description | Angel homolog 1. | |||||
|
CNOT6_HUMAN
|
||||||
| NC score | 0.372832 (rank : 4) | θ value | 1.80466e-16 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULM6 | Gene names | CNOT6, CCR4, KIAA1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
CNOT6_MOUSE
|
||||||
| NC score | 0.357457 (rank : 5) | θ value | 1.058e-16 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3P5, Q5NCL3, Q80V15 | Gene names | Cnot6, Ccr4, Kiaa1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
TTRAP_HUMAN
|
||||||
| NC score | 0.200460 (rank : 6) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95551, Q2TBE2, Q5JYM0, Q7Z6U5, Q9NUK5, Q9NYY9 | Gene names | TTRAP, EAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF and TNF receptor-associated protein (ETS1-associated protein 2) (ETS1-associated protein II) (EAPII). | |||||
|
TTRAP_MOUSE
|
||||||
| NC score | 0.159942 (rank : 7) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJX7 | Gene names | Ttrap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF and TNF receptor-associated protein. | |||||
|
CJ006_HUMAN
|
||||||
| NC score | 0.029199 (rank : 8) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IX21, Q5W0L8, Q9NPE8 | Gene names | C10orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.024052 (rank : 9) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.017572 (rank : 10) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CSTFT_HUMAN
|
||||||
| NC score | 0.016980 (rank : 11) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
CSTFT_MOUSE
|
||||||
| NC score | 0.016472 (rank : 12) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.013832 (rank : 13) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.013309 (rank : 14) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
B4GN1_HUMAN
|
||||||
| NC score | 0.012270 (rank : 15) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00973 | Gene names | B4GALNT1, GALGT, SIAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4 N-acetylgalactosaminyltransferase 1 (EC 2.4.1.92) ((N- acetylneuraminyl)-galactosylglucosylceramide) (GM2/GD2 synthase) (GalNAc-T). | |||||
|
BIG2_HUMAN
|
||||||
| NC score | 0.010804 (rank : 16) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.001247 (rank : 17) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||