Please be patient as the page loads
|
OSBL3_HUMAN
|
||||||
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OSBL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995133 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL6_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.983457 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL6_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.981870 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL7_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.982253 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
OSBP1_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 6) | NC score | 0.887020 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
OSBL1_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 7) | NC score | 0.620600 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBL1_MOUSE
|
||||||
| θ value | 1.7756e-72 (rank : 8) | NC score | 0.634199 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XL9, O88318, Q3TH97, Q673L8, Q6DFU6 | Gene names | Osbpl1a, Orp1, Orp1a | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 5.71191e-71 (rank : 9) | NC score | 0.890017 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
OSBP2_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 10) | NC score | 0.892414 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
OSBL2_HUMAN
|
||||||
| θ value | 4.68348e-65 (rank : 11) | NC score | 0.911433 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1P3, Q9BZB1, Q9Y4B8 | Gene names | OSBPL2, KIAA0772, ORP2 | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL2_MOUSE
|
||||||
| θ value | 1.15626e-55 (rank : 12) | NC score | 0.905521 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX94, Q3TCK8, Q8R0H8 | Gene names | Osbpl2 | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 13) | NC score | 0.682643 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 14) | NC score | 0.663665 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
OSBL5_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 15) | NC score | 0.681702 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
OSBL9_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 16) | NC score | 0.645827 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96SU4, Q9H9X2 | Gene names | OSBPL9, ORP9, OSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 9 (OSBP-related protein 9) (ORP-9). | |||||
|
OSB10_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 17) | NC score | 0.649243 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
OSB11_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 18) | NC score | 0.619331 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB11_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 19) | NC score | 0.608482 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BXB4 | Gene names | OSBPL11, ORP11, OSBP12 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 20) | NC score | 0.102741 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
C43BP_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 21) | NC score | 0.328384 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 22) | NC score | 0.139700 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 23) | NC score | 0.102201 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
C43BP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 24) | NC score | 0.331375 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 25) | NC score | 0.083663 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
FBX28_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.063869 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVF7, O75070 | Gene names | FBXO28, KIAA0483 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 28. | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.038679 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.029982 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.090027 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.053310 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.083231 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.083183 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.078981 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
PKHA3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.347549 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.088523 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
PKHA3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.353631 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ERS4 | Gene names | Plekha3, Fapp1 | |||
|
Domain Architecture |
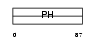
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.088847 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.013430 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.082213 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.080871 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
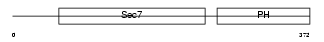
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.009045 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.013424 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.063990 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.013654 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.072017 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.020843 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.066043 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
IP3KC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.030887 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
STX12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.018369 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ER00, Q3UIV9, Q921T9 | Gene names | Stx12 | |||
|
Domain Architecture |
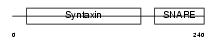
|
|||||
| Description | Syntaxin-12. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.035579 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.057829 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.010672 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.049647 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.064954 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.097130 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.048246 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.071798 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.063137 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.063538 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.009414 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.028442 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.095139 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.008578 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
IDD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.010940 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |
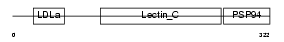
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.062730 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.024638 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SPAT7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.032721 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.017490 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.006591 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.009341 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.008107 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ST18_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.007811 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
STX12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.015276 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.011520 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.016781 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.011150 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
SIA7C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.007001 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDV1, Q6UX29, Q8N259 | Gene names | ST6GALNAC3, SIAT7C | |||
|
Domain Architecture |
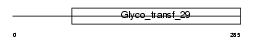
|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase III) (ST6GalNAc III) (Sialyltransferase 7C) (STY). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.013569 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K2C5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.002674 (rank : 101) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.077238 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.002882 (rank : 100) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.003988 (rank : 97) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SEM4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.003335 (rank : 98) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SLAF6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.008472 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ET39, Q9ET40 | Gene names | Slamf6, Ly108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 6 precursor (Lymphocyte antigen 108). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.069880 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
ARTS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.004893 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
K2C6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.002432 (rank : 103) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
K2C6E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.002488 (rank : 102) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.036407 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
NOP56_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.006168 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.004073 (rank : 96) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.003024 (rank : 99) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.072960 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.062043 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.052672 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
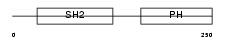
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.066296 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050587 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.068576 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.062194 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.064262 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
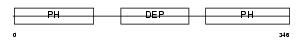
|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.065541 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
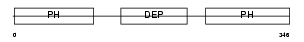
|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.052533 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.054484 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
OSBL3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL3_MOUSE
|
||||||
| NC score | 0.995133 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL6_HUMAN
|
||||||
| NC score | 0.983457 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL7_HUMAN
|
||||||
| NC score | 0.982253 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
OSBL6_MOUSE
|
||||||
| NC score | 0.981870 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL2_HUMAN
|
||||||
| NC score | 0.911433 (rank : 6) | θ value | 4.68348e-65 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1P3, Q9BZB1, Q9Y4B8 | Gene names | OSBPL2, KIAA0772, ORP2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL2_MOUSE
|
||||||
| NC score | 0.905521 (rank : 7) | θ value | 1.15626e-55 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX94, Q3TCK8, Q8R0H8 | Gene names | Osbpl2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBP2_MOUSE
|
||||||
| NC score | 0.892414 (rank : 8) | θ value | 1.66191e-70 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.890017 (rank : 9) | θ value | 5.71191e-71 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.887020 (rank : 10) | θ value | 4.67263e-73 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.682643 (rank : 11) | θ value | 5.07402e-19 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
OSBL5_MOUSE
|
||||||
| NC score | 0.681702 (rank : 12) | θ value | 1.24977e-17 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.663665 (rank : 13) | θ value | 5.60996e-18 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
OSB10_HUMAN
|
||||||
| NC score | 0.649243 (rank : 14) | θ value | 1.09485e-13 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
OSBL9_HUMAN
|
||||||
| NC score | 0.645827 (rank : 15) | θ value | 1.29331e-14 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96SU4, Q9H9X2 | Gene names | OSBPL9, ORP9, OSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 9 (OSBP-related protein 9) (ORP-9). | |||||
|
OSBL1_MOUSE
|
||||||
| NC score | 0.634199 (rank : 16) | θ value | 1.7756e-72 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XL9, O88318, Q3TH97, Q673L8, Q6DFU6 | Gene names | Osbpl1a, Orp1, Orp1a | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBL1_HUMAN
|
||||||
| NC score | 0.620600 (rank : 17) | θ value | 7.97034e-73 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSB11_MOUSE
|
||||||
| NC score | 0.619331 (rank : 18) | θ value | 1.02475e-11 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB11_HUMAN
|
||||||
| NC score | 0.608482 (rank : 19) | θ value | 5.62301e-10 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BXB4 | Gene names | OSBPL11, ORP11, OSBP12 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
PKHA3_MOUSE
|
||||||
| NC score | 0.353631 (rank : 20) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ERS4 | Gene names | Plekha3, Fapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
PKHA3_HUMAN
|
||||||
| NC score | 0.347549 (rank : 21) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
C43BP_HUMAN
|
||||||
| NC score | 0.331375 (rank : 22) | θ value | 0.00298849 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
C43BP_MOUSE
|
||||||
| NC score | 0.328384 (rank : 23) | θ value | 0.00102713 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.139700 (rank : 24) | θ value | 0.00134147 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.102741 (rank : 25) | θ value | 0.000602161 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.102201 (rank : 26) | θ value | 0.00175202 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.097130 (rank : 27) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.095139 (rank : 28) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.090027 (rank : 29) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.088847 (rank : 30) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.088523 (rank : 31) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.083663 (rank : 32) | θ value | 0.0148317 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.083231 (rank : 33) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.083183 (rank : 34) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.082213 (rank : 35) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.080871 (rank : 36) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.078981 (rank : 37) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.077238 (rank : 38) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.072960 (rank : 39) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.072017 (rank : 40) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.071798 (rank : 41) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.069880 (rank : 42) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.068576 (rank : 43) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.066296 (rank : 44) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.066043 (rank : 45) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.065541 (rank : 46) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.064954 (rank : 47) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.064262 (rank : 48) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.063990 (rank : 49) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
FBX28_HUMAN
|
||||||
| NC score | 0.063869 (rank : 50) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVF7, O75070 | Gene names | FBXO28, KIAA0483 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 28. | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.063538 (rank : 51) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.063137 (rank : 52) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.062730 (rank : 53) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.062194 (rank : 54) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.062043 (rank : 55) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.057829 (rank : 56) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.054484 (rank : 57) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.053310 (rank : 58) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.052672 (rank : 59) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.052533 (rank : 60) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.050587 (rank : 61) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.049647 (rank : 62) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.048246 (rank : 63) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.038679 (rank : 64) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.036407 (rank : 65) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.035579 (rank : 66) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SPAT7_HUMAN
|
||||||
| NC score | 0.032721 (rank : 67) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
IP3KC_MOUSE
|
||||||
| NC score | 0.030887 (rank : 68) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.029982 (rank : 69) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.028442 (rank : 70) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.024638 (rank : 71) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.020843 (rank : 72) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
STX12_MOUSE
|
||||||
| NC score | 0.018369 (rank : 73) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ER00, Q3UIV9, Q921T9 | Gene names | Stx12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.017490 (rank : 74) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.016781 (rank : 75) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
STX12_HUMAN
|
||||||
| NC score | 0.015276 (rank : 76) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.013654 (rank : 77) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.013569 (rank : 78) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.013430 (rank : 79) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.013424 (rank : 80) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.011520 (rank : 81) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.011150 (rank : 82) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
IDD_HUMAN
|
||||||
| NC score | 0.010940 (rank : 83) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.010672 (rank : 84) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.009414 (rank : 85) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.009341 (rank : 86) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.009045 (rank : 87) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.008578 (rank : 88) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SLAF6_MOUSE
|
||||||
| NC score | 0.008472 (rank : 89) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ET39, Q9ET40 | Gene names | Slamf6, Ly108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLAM family member 6 precursor (Lymphocyte antigen 108). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.008107 (rank : 90) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ST18_MOUSE
|
||||||
| NC score | 0.007811 (rank : 91) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
SIA7C_HUMAN
|
||||||
| NC score | 0.007001 (rank : 92) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDV1, Q6UX29, Q8N259 | Gene names | ST6GALNAC3, SIAT7C | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase III) (ST6GalNAc III) (Sialyltransferase 7C) (STY). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.006591 (rank : 93) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NOP56_HUMAN
|
||||||
| NC score | 0.006168 (rank : 94) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
ARTS1_HUMAN
|
||||||
| NC score | 0.004893 (rank : 95) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.004073 (rank : 96) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.003988 (rank : 97) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SEM4B_MOUSE
|
||||||
| NC score | 0.003335 (rank : 98) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.003024 (rank : 99) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.002882 (rank : 100) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
K2C5_HUMAN
|
||||||
| NC score | 0.002674 (rank : 101) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
K2C6E_HUMAN
|
||||||
| NC score | 0.002488 (rank : 102) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
K2C6B_HUMAN
|
||||||
| NC score | 0.002432 (rank : 103) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||