Please be patient as the page loads
|
OSB11_MOUSE
|
||||||
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OSB10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984326 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
OSB11_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997730 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BXB4 | Gene names | OSBPL11, ORP11, OSBP12 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB11_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSBL9_HUMAN
|
||||||
| θ value | 1.17455e-116 (rank : 4) | NC score | 0.947440 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96SU4, Q9H9X2 | Gene names | OSBPL9, ORP9, OSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 9 (OSBP-related protein 9) (ORP-9). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 2.84797e-54 (rank : 5) | NC score | 0.822209 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
OSBL5_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 6) | NC score | 0.839830 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 7) | NC score | 0.835805 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
OSBL1_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 8) | NC score | 0.489649 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBL1_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 9) | NC score | 0.502351 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91XL9, O88318, Q3TH97, Q673L8, Q6DFU6 | Gene names | Osbpl1a, Orp1, Orp1a | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
OSBP1_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 10) | NC score | 0.693366 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 11) | NC score | 0.683967 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
OSBL2_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 12) | NC score | 0.701675 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX94, Q3TCK8, Q8R0H8 | Gene names | Osbpl2 | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL2_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 13) | NC score | 0.699594 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1P3, Q9BZB1, Q9Y4B8 | Gene names | OSBPL2, KIAA0772, ORP2 | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL7_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 14) | NC score | 0.661848 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
OSBP2_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 15) | NC score | 0.661808 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
OSBL6_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 16) | NC score | 0.642938 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL3_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 17) | NC score | 0.639131 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL6_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 18) | NC score | 0.627644 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL3_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 19) | NC score | 0.619331 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
C43BP_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 20) | NC score | 0.467912 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
C43BP_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 21) | NC score | 0.463395 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
PKHA3_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 22) | NC score | 0.495263 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
PKHA3_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 23) | NC score | 0.500633 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ERS4 | Gene names | Plekha3, Fapp1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 24) | NC score | 0.135579 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 25) | NC score | 0.223944 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 26) | NC score | 0.135353 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 27) | NC score | 0.214436 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 28) | NC score | 0.094790 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 29) | NC score | 0.215710 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 30) | NC score | 0.219450 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin-2. | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 31) | NC score | 0.122145 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 32) | NC score | 0.080006 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.135472 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 34) | NC score | 0.115917 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.130541 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.117242 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.113620 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.109562 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.027559 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.045115 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.079882 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
OS9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.041762 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
CAPS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.032119 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.011298 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
CAPS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.031503 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.033044 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.044821 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.058917 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.011484 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SLIK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.010891 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H156, Q5JXB1, Q8NBC7, Q96JH3 | Gene names | SLITRK2, CXorf2, KIAA1854, SLITL1 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 2 precursor. | |||||
|
SLIK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.010836 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810C0, Q8BXL6 | Gene names | Slitrk2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 2 precursor. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.016616 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CE016_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.031401 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRJ3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16 homolog. | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.083107 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.016259 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.016353 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.014694 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.014156 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
SNX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.010767 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |
|
|||||
| Description | Sorting nexin-1. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.013201 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.014777 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ABCAD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.003917 (rank : 85) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
APS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.017439 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14492 | Gene names | APS | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
APS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.018786 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JID9, O88936 | Gene names | Aps | |||
|
Domain Architecture |
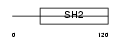
|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.005262 (rank : 82) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.004090 (rank : 84) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
K1914_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.027556 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
ODPAT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.017911 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29803 | Gene names | PDHA2 | |||
|
Domain Architecture |
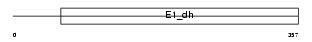
|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, testis-specific form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type II). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.004778 (rank : 83) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
K1914_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.028303 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.007980 (rank : 81) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.060392 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.058616 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
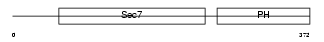
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.057564 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.057595 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056782 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.077544 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
DAPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.054296 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
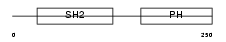
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.059375 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.073250 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.071507 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.068845 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.052491 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051350 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.062485 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
OSB11_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8CI95 | Gene names | Osbpl11 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB11_HUMAN
|
||||||
| NC score | 0.997730 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BXB4 | Gene names | OSBPL11, ORP11, OSBP12 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 11 (OSBP-related protein 11) (ORP-11). | |||||
|
OSB10_HUMAN
|
||||||
| NC score | 0.984326 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXB5, Q9BTU5 | Gene names | OSBPL10, ORP10, OSBP9 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 10 (OSBP-related protein 10) (ORP-10). | |||||
|
OSBL9_HUMAN
|
||||||
| NC score | 0.947440 (rank : 4) | θ value | 1.17455e-116 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96SU4, Q9H9X2 | Gene names | OSBPL9, ORP9, OSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 9 (OSBP-related protein 9) (ORP-9). | |||||
|
OSBL5_MOUSE
|
||||||
| NC score | 0.839830 (rank : 5) | θ value | 1.72781e-51 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.835805 (rank : 6) | θ value | 8.57503e-51 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.822209 (rank : 7) | θ value | 2.84797e-54 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
OSBL2_MOUSE
|
||||||
| NC score | 0.701675 (rank : 8) | θ value | 1.47631e-18 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BX94, Q3TCK8, Q8R0H8 | Gene names | Osbpl2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBL2_HUMAN
|
||||||
| NC score | 0.699594 (rank : 9) | θ value | 3.28887e-18 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1P3, Q9BZB1, Q9Y4B8 | Gene names | OSBPL2, KIAA0772, ORP2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 2 (OSBP-related protein 2) (ORP-2). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.693366 (rank : 10) | θ value | 2.43343e-21 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.683967 (rank : 11) | θ value | 5.07402e-19 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
OSBL7_HUMAN
|
||||||
| NC score | 0.661848 (rank : 12) | θ value | 2.7842e-17 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
OSBP2_MOUSE
|
||||||
| NC score | 0.661808 (rank : 13) | θ value | 1.80466e-16 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
OSBL6_HUMAN
|
||||||
| NC score | 0.642938 (rank : 14) | θ value | 4.44505e-15 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL3_MOUSE
|
||||||
| NC score | 0.639131 (rank : 15) | θ value | 7.58209e-15 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBS9 | Gene names | Osbpl3, Orp3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL6_MOUSE
|
||||||
| NC score | 0.627644 (rank : 16) | θ value | 3.52202e-12 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
OSBL3_HUMAN
|
||||||
| NC score | 0.619331 (rank : 17) | θ value | 1.02475e-11 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H4L5, O14591, O43357, O43358, Q8N702, Q8N703, Q8N704, Q8NFH0, Q8NFH1, Q8NI12, Q8NI13, Q9BZF4, Q9UED6 | Gene names | OSBPL3, KIAA0704, ORP3 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 3 (OSBP-related protein 3) (ORP-3). | |||||
|
OSBL1_MOUSE
|
||||||
| NC score | 0.502351 (rank : 18) | θ value | 8.93572e-24 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91XL9, O88318, Q3TH97, Q673L8, Q6DFU6 | Gene names | Osbpl1a, Orp1, Orp1a | |||
|
Domain Architecture |
|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
PKHA3_MOUSE
|
||||||
| NC score | 0.500633 (rank : 19) | θ value | 1.99992e-07 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ERS4 | Gene names | Plekha3, Fapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
PKHA3_HUMAN
|
||||||
| NC score | 0.495263 (rank : 20) | θ value | 8.97725e-08 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
OSBL1_HUMAN
|
||||||
| NC score | 0.489649 (rank : 21) | θ value | 1.80048e-24 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
C43BP_HUMAN
|
||||||
| NC score | 0.467912 (rank : 22) | θ value | 1.47974e-10 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
C43BP_MOUSE
|
||||||
| NC score | 0.463395 (rank : 23) | θ value | 1.9326e-10 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.223944 (rank : 24) | θ value | 0.000121331 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
PLEK2_MOUSE
|
||||||
| NC score | 0.219450 (rank : 25) | θ value | 0.00509761 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WV52 | Gene names | Plek2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.215710 (rank : 26) | θ value | 0.00509761 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.214436 (rank : 27) | θ value | 0.00035302 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.135579 (rank : 28) | θ value | 9.29e-05 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.135472 (rank : 29) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.135353 (rank : 30) | θ value | 0.00020696 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.130541 (rank : 31) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.122145 (rank : 32) | θ value | 0.0148317 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.117242 (rank : 33) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.115917 (rank : 34) | θ value | 0.0330416 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.113620 (rank : 35) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.109562 (rank : 36) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.094790 (rank : 37) | θ value | 0.00298849 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.083107 (rank : 38) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.080006 (rank : 39) | θ value | 0.0252991 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.079882 (rank : 40) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.077544 (rank : 41) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.073250 (rank : 42) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.071507 (rank : 43) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.068845 (rank : 44) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.062485 (rank : 45) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.060392 (rank : 46) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.059375 (rank : 47) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.058917 (rank : 48) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.058616 (rank : 49) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.057595 (rank : 50) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.057564 (rank : 51) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.056782 (rank : 52) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.054296 (rank : 53) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.052491 (rank : 54) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.051350 (rank : 55) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.045115 (rank : 56) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.044821 (rank : 57) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.041762 (rank : 58) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.033044 (rank : 59) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
CAPS1_HUMAN
|
||||||
| NC score | 0.032119 (rank : 60) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_MOUSE
|
||||||
| NC score | 0.031503 (rank : 61) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CE016_MOUSE
|
||||||
| NC score | 0.031401 (rank : 62) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRJ3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16 homolog. | |||||
|
K1914_MOUSE
|
||||||
| NC score | 0.028303 (rank : 63) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.027559 (rank : 64) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
K1914_HUMAN
|
||||||
| NC score | 0.027556 (rank : 65) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
APS_MOUSE
|
||||||
| NC score | 0.018786 (rank : 66) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JID9, O88936 | Gene names | Aps | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
ODPAT_HUMAN
|
||||||
| NC score | 0.017911 (rank : 67) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29803 | Gene names | PDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase E1 component alpha subunit, testis-specific form, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A type II). | |||||
|
APS_HUMAN
|
||||||
| NC score | 0.017439 (rank : 68) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14492 | Gene names | APS | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.016616 (rank : 69) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.016353 (rank : 70) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.016259 (rank : 71) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.014777 (rank : 72) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.014694 (rank : 73) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.014156 (rank : 74) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.013201 (rank : 75) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.011484 (rank : 76) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.011298 (rank : 77) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SLIK2_HUMAN
|
||||||
| NC score | 0.010891 (rank : 78) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H156, Q5JXB1, Q8NBC7, Q96JH3 | Gene names | SLITRK2, CXorf2, KIAA1854, SLITL1 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 2 precursor. | |||||
|
SLIK2_MOUSE
|
||||||
| NC score | 0.010836 (rank : 79) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810C0, Q8BXL6 | Gene names | Slitrk2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 2 precursor. | |||||
|
SNX1_MOUSE
|
||||||
| NC score | 0.010767 (rank : 80) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-1. | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.007980 (rank : 81) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.005262 (rank : 82) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.004778 (rank : 83) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.004090 (rank : 84) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
ABCAD_MOUSE
|
||||||
| NC score | 0.003917 (rank : 85) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||